8AHI
 
 | |
1ULW
 
 | | Crystal structure of P450nor Ser73Gly/Ser75Gly mutant | | Descriptor: | Cytochrome P450 55A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Su, F, Li, Z, Takaya, N, Shoun, H. | | Deposit date: | 2003-09-16 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
3ROK
 
 | |
1XQD
 
 | | Crystal structure of P450NOR complexed with 3-pyridinealdehyde adenine dinucleotide | | Descriptor: | CYTOCHROME P450 55A1, NICOTINIC ACID ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Takaya, N, Su, F, Wakagi, T, Shoun, H. | | Deposit date: | 2004-10-12 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
3ROQ
 
 | | Crystal structure of human CD38 in complex with compound CZ-46 | | Descriptor: | (2R,3R,4S,5S)-4-fluoro-3,5-dihydroxytetrahydrofuran-2-yl 2-phenylethyl hydrogen (S)-phosphate, ADP-ribosyl cyclase 1 | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-04-26 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalysis-based inhibitors of the calcium signaling function of CD38.
Biochemistry, 51, 2012
|
|
3ROM
 
 | |
3NCG
 
 | |
3NYV
 
 | |
6ZI9
 
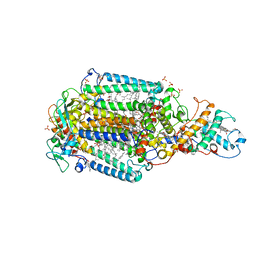 | | Ultrafast Structural Response to Charge Redistribution Within a Photosynthetic Reaction Centre - 300 ps (b) structure | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baath, P, Dods, R, Braenden, G, Neutze, R. | | Deposit date: | 2020-06-25 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ultrafast structural changes within a photosynthetic reaction centre.
Nature, 589, 2021
|
|
7X2A
 
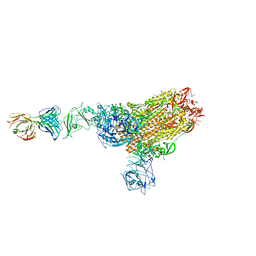 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class1 (1u2d RBD with 1Fab) | | Descriptor: | MERS-CoV Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X26
 
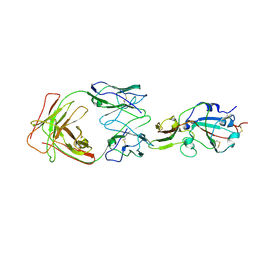 | | S41 neutralizing antibody Fab(MERS-CoV) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.685 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X29
 
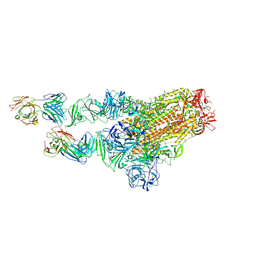 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class2 (1u2d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X28
 
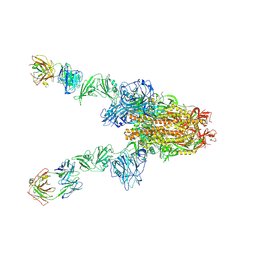 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class3 (2u1d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
6ZID
 
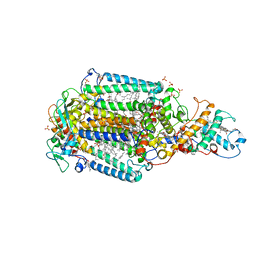 | | Ultrafast Structural Response to Charge Redistribution Within a Photosynthetic Reaction Centre - 5 ps (b) structure | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baath, P, Dods, R, Braenden, G, Neutze, R. | | Deposit date: | 2020-06-25 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ultrafast structural changes within a photosynthetic reaction centre.
Nature, 589, 2021
|
|
6ZI4
 
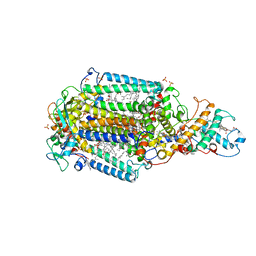 | | Ultrafast Structural Response to Charge Redistribution Within a Photosynthetic Reaction Centre - 5 ps (a) structure | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baath, P, Dods, R, Braenden, G, Neutze, R. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ultrafast structural changes within a photosynthetic reaction centre.
Nature, 589, 2021
|
|
4ETB
 
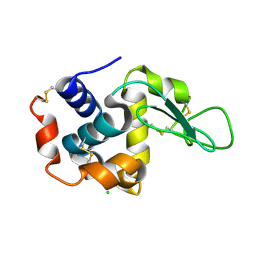 | | lysozyme, room temperature, 200 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
6ZI5
 
 | | Ultrafast Structural Response to Charge Redistribution Within a Photosynthetic Reaction Centre - 300 ps (a) structure | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baath, P, Dods, R, Braenden, G, Neutze, R. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ultrafast structural changes within a photosynthetic reaction centre.
Nature, 589, 2021
|
|
5NHH
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-2-[2-(oxan-4-ylamino)pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHO
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | (6~{S})-5-(2-methoxyethyl)-6-methyl-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
6ZIA
 
 | | Ultrafast Structural Response to Charge Redistribution Within a Photosynthetic Reaction Centre - 8 us structure | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baath, P, Dods, R, Braenden, G, Neutze, R. | | Deposit date: | 2020-06-25 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ultrafast structural changes within a photosynthetic reaction centre.
Nature, 589, 2021
|
|
5NGU
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 2-[2-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHV
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 7-[2-(oxan-4-ylamino)pyrimidin-4-yl]-3,4-dihydro-2~{H}-pyrrolo[1,2-a]pyrazin-1-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
4ETE
 
 | | Lysozyme, room-temperature, rotating anode, 0.0021 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
7CCE
 
 | |
