5EHZ
 
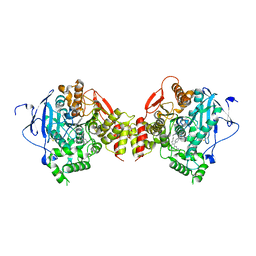 | | mAChE-syn TZ2PA5 complex from an equimolar mixture of the syn/anti isomers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-phenyl-5-[5-[3-[2-(1,2,3,4-tetrahydroacridin-9-ylamino)ethyl]-1,2,3-triazol-4-yl]pentyl]phenanthridin-5-ium-3,8-diamine, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
5EIA
 
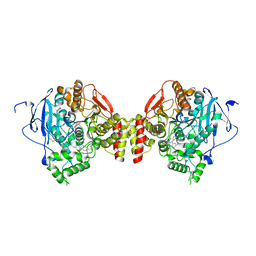 | | mACHE-anti TZ2PA5 complex from a 1:6 mixture of the syn/anti isomers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-phenyl-5-[5-[1-[2-(1,2,3,4-tetrahydroacridin-9-ylamino)ethyl]-1,2,3-triazol-4-yl]pentyl]phenanthridin-5-ium-3,8-diamine, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
5EIE
 
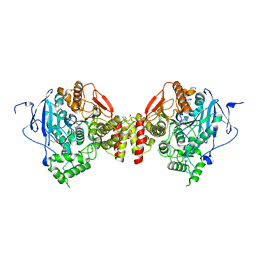 | | mAChE-TZ2 complex | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
5EHQ
 
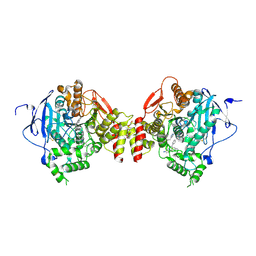 | | mAChE-anti TZ2PA5 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 6-phenyl-5-[5-[1-[2-(1,2,3,4-tetrahydroacridin-9-ylamino)ethyl]-1,2,3-triazol-4-yl]pentyl]phenanthridin-5-ium-3,8-diamine, ... | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
6O50
 
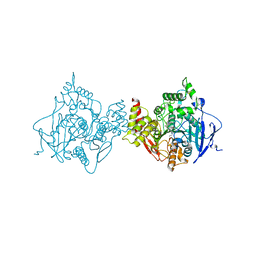 | | Binary complex of native hAChE with BW284c51 | | Descriptor: | 4-(5-{4-[DIMETHYL(PROP-2-ENYL)AMMONIO]PHENYL}-3-OXOPENTYL)-N,N-DIMETHYL-N-PROP-2-ENYLBENZENAMINIUM, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies.
Chem.Biol.Interact., 309, 2019
|
|
6O4X
 
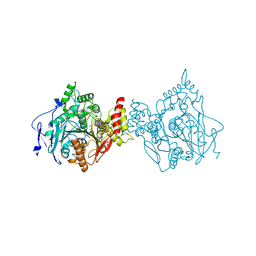 | | Binary complex of native hAChE with 9-aminoacridine | | Descriptor: | 9-AMINOACRIDINE, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies.
Chem.Biol.Interact., 309, 2019
|
|
6O5S
 
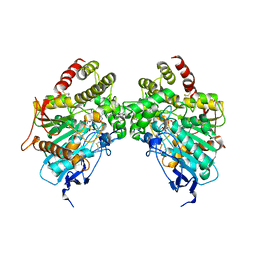 | |
6O5V
 
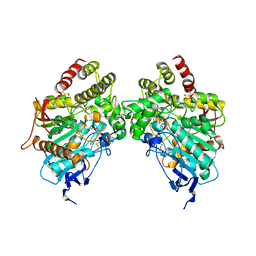 | | Binary complex of native hAChE with oxime reactivator RS-170B | | Descriptor: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-04 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Productive reorientation of a bound oxime reactivator revealed in room temperature X-ray structures of native and VX-inhibited human acetylcholinesterase.
J.Biol.Chem., 294, 2019
|
|
6O66
 
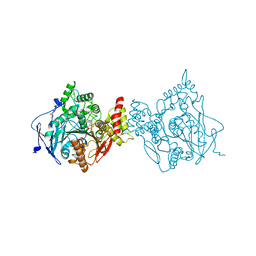 | |
6O4W
 
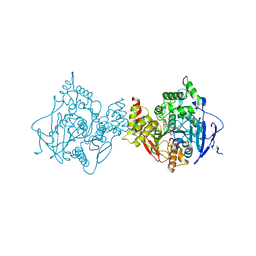 | | Binary complex of native hAChE with Donepezil | | Descriptor: | 1-BENZYL-4-[(5,6-DIMETHOXY-1-INDANON-2-YL)METHYL]PIPERIDINE, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies.
Chem.Biol.Interact., 309, 2019
|
|
6O52
 
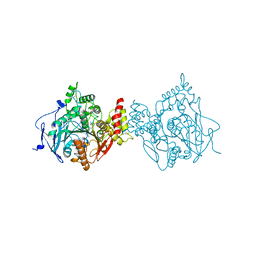 | |
6O5R
 
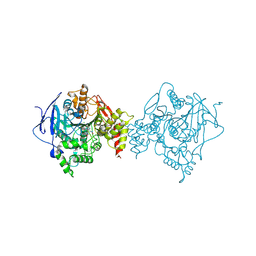 | |
4YF5
 
 | |
4YF6
 
 | | Crystal structure of oxidised Rv1284 | | Descriptor: | Beta-carbonic anhydrase 1, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hofmann, A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Chemical probing suggests redox-regulation of the carbonic anhydrase activity of mycobacterial Rv1284.
Febs J., 282, 2015
|
|
4YF4
 
 | |
4NUI
 
 | | Crystal structure of cobalt-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, COBALT (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
2IGU
 
 | | Deamidated analogue of ImI Conotoxin | | Descriptor: | Alpha-conotoxin ImI | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-25 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
4NUO
 
 | | Crystal structure of zinc-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, ZINC ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
4NUK
 
 | | Crystal structure of nickel-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, NICKEL (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
2IH6
 
 | | Pro6 variant of CMrVIA conotoxin | | Descriptor: | Lambda-conotoxin CMrVIA | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IHA
 
 | | Amidated variant of CMrVIA conotoxin | | Descriptor: | Lambda-conotoxin CMrVIA | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IFZ
 
 | | Lys6 Variant of ImI Conotoxin | | Descriptor: | Alpha-conotoxin ImI | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-22 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IFI
 
 | | Ala6 Variant of ImI Conotoxin | | Descriptor: | Alpha-conotoxin ImI | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-21 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IH7
 
 | | Amidated Pro6 Analogue of CMrVIA conotoxin | | Descriptor: | Lambda-conotoxin CMrVIA | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IFJ
 
 | | Lys6 deamidated variant of ImI conotoxin | | Descriptor: | Alpha-conotoxin ImI | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-21 | | Release date: | 2007-08-14 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
