7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
8AZK
 
 | | Bovine 20S proteasome, untreated | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Szenkier, N, Arie, M, Matzov, D, Sertchook, R, Carmeli, R, Cascio, P, Stanhill, A, Shalev Benami, M, Navon, A. | | Deposit date: | 2022-09-06 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bovine 20S proteasome, untreated
To be published
|
|
5YWX
 
 | | Crystal structure of hematopoietic prostaglandin D synthase in complex with F092 | | Descriptor: | GLUTATHIONE, GLYCEROL, Hematopoietic prostaglandin D synthase, ... | | Authors: | Kamo, M, Furubayashi, N, Inaka, K, Aritake, K, Omura, A, Tanaka, A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Characterization of crystal water molecules in a high-affinity inhibitor and hematopoietic prostaglandin D synthase complex by interaction energy studies.
Bioorg. Med. Chem., 26, 2018
|
|
8B29
 
 | | Human carbonic anhydrase II containing 6-fluorotryptophanes. | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2022-09-13 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Direct Expression of Fluorinated Proteins in Human Cells for 19 F In-Cell NMR Spectroscopy.
J.Am.Chem.Soc., 145, 2023
|
|
4V26
 
 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-CHLORO-5-METHYLPYRIMIDIN-4-YL)PHENYL]-2,4-DIHYDROXY-N-(4-{[(TRIFLUOROACETYL)AMINO]METHYL}BENZYL)BENZAMIDE, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
8KCQ
 
 | | Solution structures of the N-terminal divergent caplonin homology (NN-CH) domains of human intraflagellar transport protein 54 | | Descriptor: | TRAF3-interacting protein 1 | | Authors: | Dang, W, Kuwasako, K, He, F, Takahashi, M, Tsuda, K, Nagata, T, Tanaka, A, Kobayashi, N, Kigawa, T, Guentert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2023-08-08 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structure of the N-terminal divergent calponin homology (NN-CH) domain of human intraflagellar transport protein 54.
Biomol.Nmr Assign., 18, 2024
|
|
3W88
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-200 | | Descriptor: | 1,2-ETHANEDIOL, 5-[4-(6-carboxynaphthalen-2-yl)butyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, CACODYLATE ION, ... | | Authors: | Inaoka, D.K, Hashimoto, S, Rocha, J.R, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Kuranaga, T, Shiba, T, Balogun, E.O, Sakamoto, K, Suzuki, S, Montanari, C.A, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-200
To be Published
|
|
8RFB
 
 | | Cryo-EM structure of the R243C mutant of human Prolyl Endopeptidase-Like (PREPL) protein involved in Congenital myasthenic syndrome-22 (CMS22) | | Descriptor: | Prolyl endopeptidase-like | | Authors: | Theodoropoulou, A, Cavani, E, Antanasijevic, A, Marcaida, M.J, Dal Peraro, M. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Missense variants in CMS22 patients reveal that PREPL has both enzymatic and nonenzymatic functions.
JCI Insight, 9, 2024
|
|
7Q64
 
 | | Cryo-em structure of the Nup98 fibril polymorph 1 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
7Q65
 
 | | Cryo-em structure of the Nup98 fibril polymorph 2 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
7Q67
 
 | | Cryo-em structure of the Nup98 fibril polymorph 4 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
7Q66
 
 | | Cryo-em structure of the Nup98 fibril polymorph 3 | | Descriptor: | Nuclear pore complex protein Nup98 | | Authors: | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
3W86
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-96 | | Descriptor: | 1,2-ETHANEDIOL, 5-{4-[4-(methoxycarbonyl)phenyl]butyl}-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, CACODYLATE ION, ... | | Authors: | Inaoka, D.K, Hashimoto, S, Rocha, J.R, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Kuranaga, T, Shiba, T, Balogun, E.O, Sakamoto, K, Suzuki, S, Montanari, C.A, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-96
To be Published
|
|
3W87
 
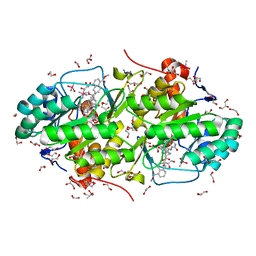 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-103 | | Descriptor: | 1,2-ETHANEDIOL, 5-{4-[5-(methoxycarbonyl)naphthalen-2-yl]butyl}-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, CACODYLATE ION, ... | | Authors: | Inaoka, D.K, Hashimoto, S, Rocha, J.R, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Kuranaga, T, Shiba, T, Balogun, E.O, Sakamoto, K, Suzuki, S, Montanari, C.A, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-103
To be Published
|
|
8PQX
 
 | | p97 (VCP) mutant - F539A state III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2023-07-12 | | Release date: | 2024-05-29 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A non-symmetrical p97 conformation initiates a multistep recruitment of Ufd1/Npl4.
Iscience, 27, 2024
|
|
1WQU
 
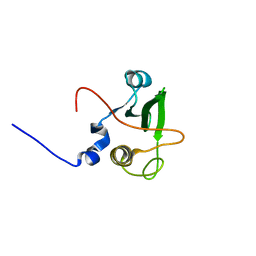 | | Solution structure of the human FES SH2 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase FES/FPS | | Authors: | Scott, A, Pantoja-Uceda, D, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Tanaka, A, Sugano, S, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-02 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Src homology 2 domain from the human feline sarcoma oncogene Fes
J.Biomol.NMR, 31, 2005
|
|
8R0E
 
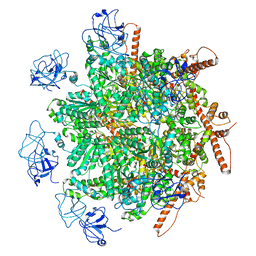 | | p97 (VCP) mutant - F266A | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-29 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A non-symmetrical p97 conformation initiates a multistep recruitment of Ufd1/Npl4.
Iscience, 27, 2024
|
|
8RGM
 
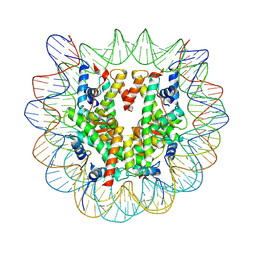 | | Cryo-EM structure of nucleosome containing Widom603 DNA | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Motorin, N.A, Afonin, D, Armeev, G.A, Moiseenko, A, Zhao, L, Vasiliev, V, Oleinikov, P, Shaytan, A, Shi, X, Studitsky, V, Sokolova, O. | | Deposit date: | 2023-12-14 | | Release date: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and unwrapping states of the Widom 603 based nucleosome.
To Be Published
|
|
8RS9
 
 | | p97 (VCP) double mutant - F266A F539A | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A non-symmetrical p97 conformation initiates a multistep recruitment of Ufd1/Npl4.
Iscience, 27, 2024
|
|
8RSB
 
 | | p97 (VCP) mutant - F539A ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A non-symmetrical p97 conformation initiates a multistep recruitment of Ufd1/Npl4.
Iscience, 27, 2024
|
|
8RSC
 
 | | p97 (VCP) mutant - F539A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A non-symmetrical p97 conformation initiates a multistep recruitment of Ufd1/Npl4.
Iscience, 27, 2024
|
|
5GN7
 
 | | Crystal structure of alternative oxidase from Trypanosoma brucei brucei complexed with cumarin derivative-17 | | Descriptor: | 4-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-7,8-bis(oxidanyl)chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5GN9
 
 | | Crystal structure of alternative oxidase from Trypanosoma brucei brucei complexed with cumarin derivative-17b | | Descriptor: | 4-butyl-7,8-bis(oxidanyl)chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5GN5
 
 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17 | | Descriptor: | 4-[[4-(4-methoxyphenyl)piperazin-1-yl]methyl]-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
7SBX
 
 | | Structure of OC43 spike in complex with polyclonal Fab6 (Donor 1051) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
