5DIC
 
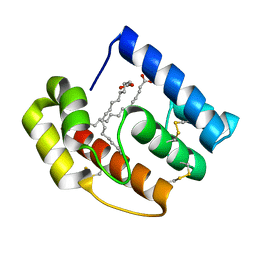 | |
1YGS
 
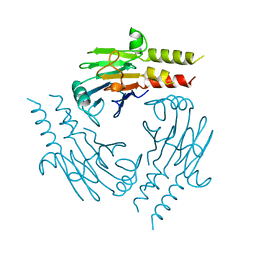 | | CRYSTAL STRUCTURE OF THE SMAD4 TUMOR SUPPRESSOR C-TERMINAL DOMAIN | | Descriptor: | SMAD4 | | Authors: | Shi, Y, Hata, A, Lo, R.S, Massague, J, Pavletich, N.P. | | Deposit date: | 1997-10-03 | | Release date: | 1998-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for mutational inactivation of the tumour suppressor Smad4.
Nature, 388, 1997
|
|
5ZQU
 
 | | Crystal structure of tetrameric RXRalpha-LBD complexed with partial agonist CBt-PMN | | Descriptor: | 1-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)benzotriazole-5-carboxylic acid, BROMIDE ION, Retinoic acid receptor RXR-alpha | | Authors: | Miyashita, Y, Numoto, N, Arulmozhiraja, S, Nakano, S, Matsuo, N, Shimizu, K, Kakuta, H, Ito, S, Ikura, T, Ito, N, Tokiwa, H. | | Deposit date: | 2018-04-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.60038781 Å) | | Cite: | Dual conformation of the ligand induces the partial agonistic activity of retinoid X receptor alpha (RXR alpha ).
FEBS Lett., 593, 2019
|
|
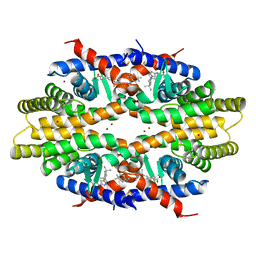
1JDC
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 1) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1JDD
 
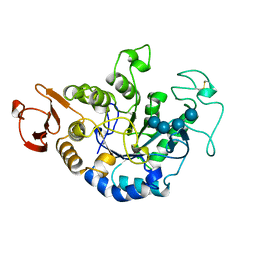 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 2) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
2RSE
 
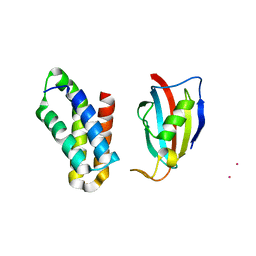 | | NMR structure of FKBP12-mTOR FRB domain-rapamycin complex structure determined based on PCS | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Serine/threonine-protein kinase mTOR, TERBIUM(III) ION | | Authors: | Kobashigawa, Y, Ushio, M, Saio, T, Inagaki, F. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination.
J.Biomol.Nmr, 53, 2012
|
|
1JDA
 
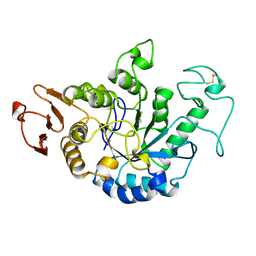 | | MALTOTETRAOSE-FORMING EXO-AMYLASE | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1IZ0
 
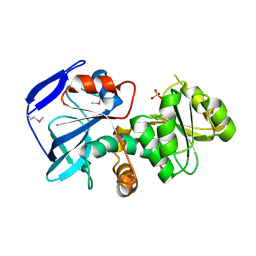 | |
1IYZ
 
 | |
1BYV
 
 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CALCITONIN) | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K.G, Opella, S.J. | | Deposit date: | 1998-10-16 | | Release date: | 1998-10-28 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
1BZB
 
 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | PROTEIN (CALCITONIN), alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-10-27 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
6AJN
 
 | | Crystal structure of AtaTR bound with AcCoA | | Descriptor: | ACETYL COENZYME *A, DUF1778 domain-containing protein, N-acetyltransferase | | Authors: | Yashiro, Y, Yamashita, S, Tomita, K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal Structure of the Enterohemorrhagic Escherichia coli AtaT-AtaR Toxin-Antitoxin Complex.
Structure, 27, 2019
|
|
1BKU
 
 | | EFFECTS OF GLYCOSYLATION ON THE STRUCTURE AND DYNAMICS OF EEL CALCITONIN, NMR, 10 STRUCTURES | | Descriptor: | CALCITONIN | | Authors: | Hashimoto, Y, Nishikido, J, Toma, K, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-07-13 | | Release date: | 1999-01-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
6AJM
 
 | | Crystal structure of apo AtaTR | | Descriptor: | DUF1778 domain-containing protein, N-acetyltransferase, TRIETHYLENE GLYCOL | | Authors: | Yashiro, Y, Yamashita, S, Tomita, K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal Structure of the Enterohemorrhagic Escherichia coli AtaT-AtaR Toxin-Antitoxin Complex.
Structure, 27, 2019
|
|
2EYY
 
 | | CT10-Regulated Kinase isoform I | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
7CT4
 
 | | Crystal structure of D-amino acid oxidase from Rasamsonia emersonii strain YA | | Descriptor: | D-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Shimekake, Y, Hirato, Y, Okazaki, S, Funabashi, R, Goto, M, Furuichi, T, Suzuki, H, Takahashi, S. | | Deposit date: | 2020-08-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of a unique D-amino-acid oxidase from the thermophilic fungus Rasamsonia emersonii strain YA.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8GQ9
 
 | | Crystal structure of lasso peptide epimerase MslH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-08-29 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of lasso peptide epimerase MslH reveals metal-dependent acid/base catalytic mechanism.
Nat Commun, 14, 2023
|
|
8GQB
 
 | |
8GQA
 
 | |
8ITH
 
 | | Crystal structure of lasso peptide epimerase MslH H295N | | Descriptor: | CALCIUM ION, GLYCEROL, Poly-gamma-glutamate synthesis protein (Capsule biosynthesis protein) | | Authors: | Nakashima, Y, Hiroyuki, M. | | Deposit date: | 2023-03-22 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of lasso peptide epimerase MslH reveals metal-dependent acid/base catalytic mechanism.
Nat Commun, 14, 2023
|
|
8ITG
 
 | |
8H8N
 
 | | Crystal structure of apo-R52Y/E56Y/R59Y/E63Y-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8L
 
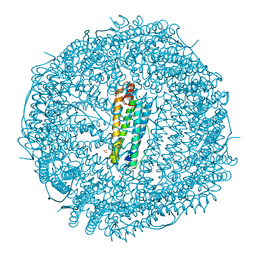 | | Crystal structure of apo-R52F/E56F/R59F/E63F-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8M
 
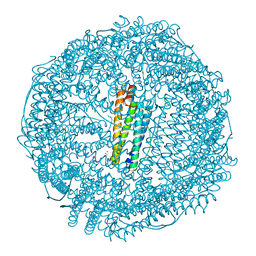 | | Crystal structure of apo-E53F/E57F/E60F/E64F-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8O
 
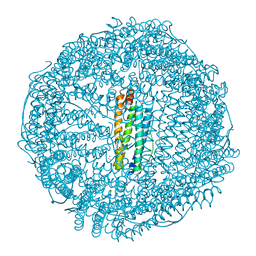 | | Crystal structure of apo-R52W/E56W/R59W/E63W-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
