5HW2
 
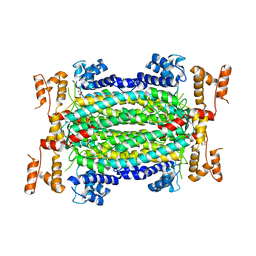 | | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid | | Descriptor: | 1,2-ETHANEDIOL, Adenylosuccinate lyase, FUMARIC ACID, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid
To Be Published
|
|
5HJ5
 
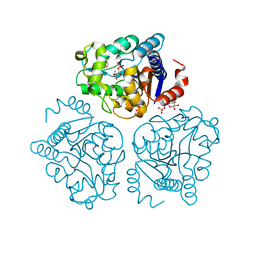 | | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, ACETIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE -6-PHOSPHATE
To Be Published
|
|
6NIO
 
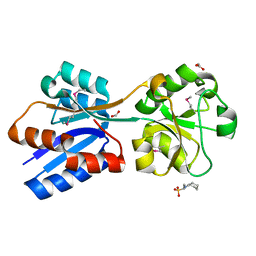 | | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ACETIC ACID, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, G, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis
To Be Published
|
|
6NI1
 
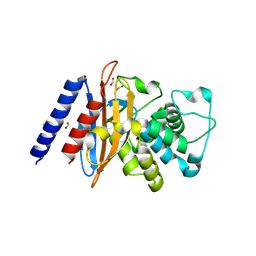 | | Crystal Structure of the Beta Lactamase Class A penP from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-25 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class A penP from Bacillus subtilis
To Be Published
|
|
6NPO
 
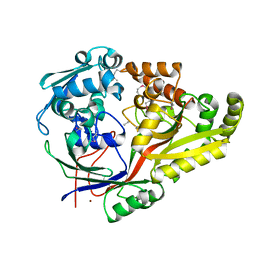 | | Crystal structure of oligopeptide ABC transporter from Bacillus anthracis str. Ames (substrate-binding domain) | | Descriptor: | Oligopeptide ABC transporter, oligopeptide-binding protein, Unknown peptide ligand, ... | | Authors: | Michalska, K, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of oligopeptide ABC transporter from
Bacillus anthracis str. Ames (substrate-binding domain)
To Be Published
|
|
6X4I
 
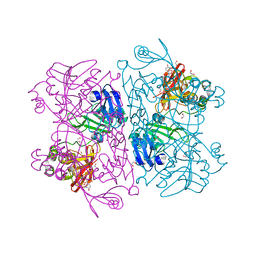 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with 3'-uridinemonophosphate | | Descriptor: | 1,2-ETHANEDIOL, 3'-URIDINEMONOPHOSPHATE, SODIUM ION, ... | | Authors: | Chang, C, Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6X1B
 
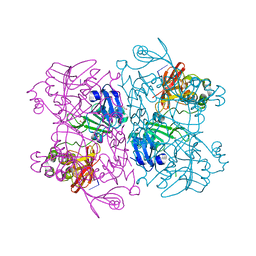 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with the Product Nucleotide GpU. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-R(*GP*U)-3'), PHOSPHATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-18 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
5HG0
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM | | Descriptor: | Pantothenate synthetase, S-ADENOSYLMETHIONINE | | Authors: | Chang, C, Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-07 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM
To Be Published
|
|
5HI6
 
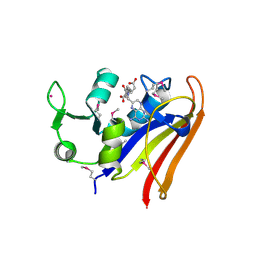 | | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form
To Be Published
|
|
5HD6
 
 | | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, GLYCEROL | | Authors: | Chang, C, Maltseva, N, Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A
To Be Published
|
|
6WXC
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with potential repurposing drug Tipiracil | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
5FDA
 
 | | The high resolution structure of apo form dihydrofolate reductase from Yersinia pestis at 1.55 A | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | structure of dihydrofolate reductase from Yersinia pestis complex with
To Be Published
|
|
4IX2
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, complexed with IMP | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, POTASSIUM ION | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, complexed with IMP.
To be Published
|
|
4IQE
 
 | | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis str. Ames Ancestor | | Descriptor: | Carboxyvinyl-carboxyphosphonate phosphorylmutase | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
4IQD
 
 | | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis | | Descriptor: | Carboxyvinyl-carboxyphosphonate phosphorylmutase, PYRUVIC ACID | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis
To be Published
|
|
8VLC
 
 | | Crystal structure of Zn-dependent hydrolase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, HARLDQ motif MBL-fold protein, SULFATE ION | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-01-11 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Zn-dependent hydrolase from Salmonella typhimurium LT2
To Be Published
|
|
6XFS
 
 | | Class C beta-lactamase from Escherichia coli in complex with Tazobactam | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Class C beta-lactamase from Escherichia coli in complex with Tazobactam
To Be Published
|
|
6XJ3
 
 | | Crystal structure of Class D beta-lactamase from Klebsiella quasipneumoniae in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Class D beta-lactamase from Klebsiella quasipneumoniae
To Be Published
|
|
6XG1
 
 | | Class C beta-lactamase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Class C beta-lactamase from Escherichia coli
To Be Published
|
|
7T8O
 
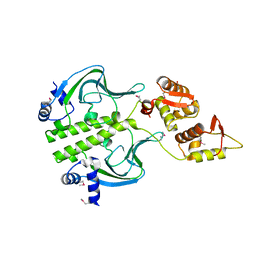 | | Crystal Structure of the Crp/Fnr Family Transcriptional Regulator from Listeria monocytogenes | | Descriptor: | Lmo0753 protein, SULFATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Shatsman, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-16 | | Release date: | 2021-12-29 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of the Crp/Fnr Family Transcriptional Regulator from Listeria monocytogenes
To Be Published
|
|
7TAV
 
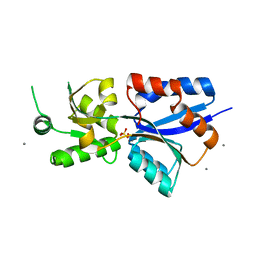 | | Crystal Structure of the PBP2_YvgL_like protein Lmo1041 from Listeria monocytogene | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of the PBP2_YvgL_like protein Lmo1041 from Listeria monocytogenes
To Be Published
|
|
7TCB
 
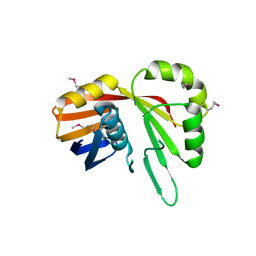 | | Crystal Structure of the YaeQ Family Protein VPA0551 from Vibrio parahaemolyticus | | Descriptor: | YaeQ family protein VPA0551 | | Authors: | Kim, Y, Mulligan, R, Maltseva, N, Gu, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-05 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the YaeQ Family Protein VPA0551 from Vibrio parahaemolyticus
To Be Published
|
|
7STS
 
 | | Crystal Structure of Human Fab S24-1379 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | Fab S24-1379, heavy chain, light chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7SUE
 
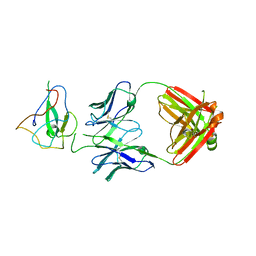 | | Crystal Structure of Human Fab S24-188 in the complex with the N-teminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | Nucleoprotein, S24-188 Fab Heavy chain, S24-188 Fab Light chain | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7STR
 
 | | Crystal Structure of Human Fab S24-1063 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Fab S24-1063, Heavy chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
