7OXO
 
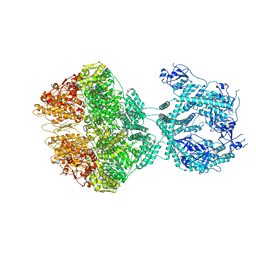 | | human LonP1, R-state, incubated in AMPPCP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Abrahams, J.P, Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
6G2R
 
 | | Crystal structure of FimH in complex with a tetraflourinated biphenyl alpha D-mannoside | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[3-chloranyl-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-phenyl]-2,3,5,6-tetrakis(fluoranyl)benzenecarbonitrile, SULFATE ION, ... | | Authors: | Jakob, R.P, Schoenemann, W, Cramer, J, Muehlethaler, T, Daetwyler, P, Zihlmann, P, Fiege, B, Sager, C.P, Smiesko, M, Rabbani, S, Eris, D, Schwardt, O, Maier, T, Ernst, B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Chemmedchem, 14, 2019
|
|
6G2S
 
 | | Crystal structure of FimH in complex with a pentaflourinated biphenyl alpha D-mannoside | | Descriptor: | (2~{R},3~{S},4~{S},5~{S},6~{R})-2-(hydroxymethyl)-6-[4-[2,3,4,5,6-pentakis(fluoranyl)phenyl]phenoxy]oxane-3,4,5-triol, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin | | Authors: | Jakob, R.P, Schoenemann, W, Cramer, J, Muehlethaler, T, Daetwyler, P, Zihlmann, P, Fiege, B, Sager, C.P, Smiesko, M, Rabbani, S, Eris, D, Schwardt, O, Maier, T, Ernst, B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Chemmedchem, 14, 2019
|
|
7P1C
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin B | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein assembly factor BamA, TRP-ASN-UX8-THR-LYS-ARG-PHE | | Authors: | Jakob, R.P, Modaresi, S.M, Hiller, S, Maier, T. | | Deposit date: | 2021-07-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutasynthetic Production and Antimicrobial Characterization of Darobactin Analogs.
Microbiol Spectr, 9, 2021
|
|
6GU0
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 with bound dimannoside Man(alpha1-3)Man in space group P213 | | Descriptor: | FimG protein, FimH protein, SULFATE ION, ... | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
6GTY
 
 | | Crystal structure of the FimH lectin domain from E.coli K12 in complex with the dimannoside Man(alpha1-6)Man | | Descriptor: | Type 1 fimbrin D-mannose specific adhesin, alpha-D-mannopyranose-(1-6)-methyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
6GTX
 
 | | Crystal structure of the FimH lectin domain from E.coli K12 in complex with the dimannoside Man(alpha1-2)Man | | Descriptor: | SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
7PE9
 
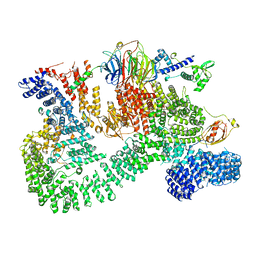 | |
7PEB
 
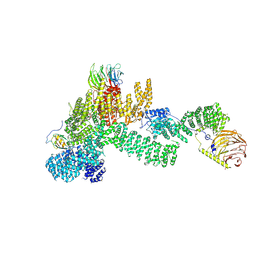 | |
7PE8
 
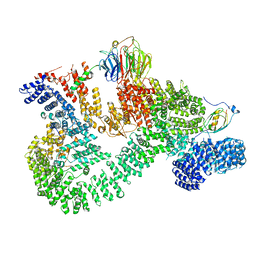 | |
7PEC
 
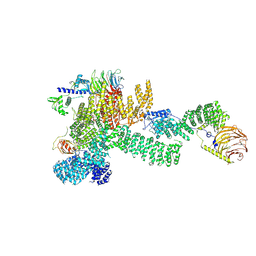 | |
7PEA
 
 | |
7PED
 
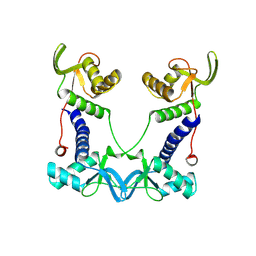 | | DEPTOR DEP domain tandem (DEPt) | | Descriptor: | DEP domain-containing mTOR-interacting protein | | Authors: | Waelchli, M, Jakob, R, Maier, T. | | Deposit date: | 2021-08-09 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Regulation of human mTOR complexes by DEPTOR.
Elife, 10, 2021
|
|
7PE7
 
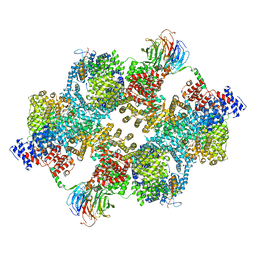 | |
6GTW
 
 | | Crystal structure of the FimH lectin domain from E.coli F18 in complex with trimannose | | Descriptor: | CALCIUM ION, FimH protein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
6GTZ
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 with bound dimannoside Man(alpha1-3)Man in space group P21 | | Descriptor: | FimG protein, FimH protein, alpha-D-mannopyranose-(1-3)-methyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
6GTV
 
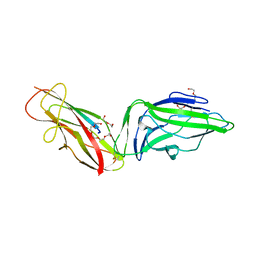 | | Crystal structure of a FimH*DsG complex from E.coli F18 with bound trimannose | | Descriptor: | DI(HYDROXYETHYL)ETHER, FimG protein, FimH protein, ... | | Authors: | Jakob, R.P, Sauer, M.M, Luber, T, Canonica, F, Navarra, G, Ernst, B, Unverzagt, C, Maier, T, Glockshuber, R. | | Deposit date: | 2018-06-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of the Bacterial Adhesin FimH to Its Natural, Multivalent High-Mannose Type Glycan Targets.
J.Am.Chem.Soc., 141, 2019
|
|
6HP1
 
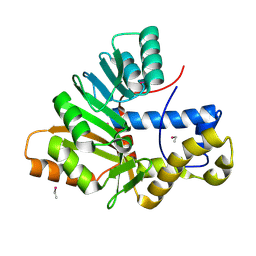 | | Crystal Structure of the O-Methyltransferase from the trans-AT PKS multienzyme C0ZGQ3 of Brevibacillus brevis | | Descriptor: | ETHYL MERCURY ION, Putative polyketide synthase | | Authors: | Jakob, R.P, Felber, P, Demyanenko, Y, Delbart, F, Maier, T. | | Deposit date: | 2018-09-19 | | Release date: | 2019-10-09 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of an O-Methyltransferase from the trans-AT PKS biosynthesis pathway
To Be Published
|
|
5MPT
 
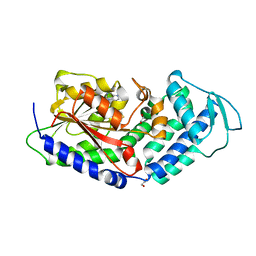 | | Structure of the citrinin polyketide synthase CMeT domain | | Descriptor: | 1,2-ETHANEDIOL, Citrinin polyketide synthase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Herbst, D.A, Storm, P.A, Townsend, C.A, Maier, T. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Functional and Structural Analysis of Programmed C-Methylation in the Biosynthesis of the Fungal Polyketide Citrinin.
Cell Chem Biol, 24, 2017
|
|
5MCA
 
 | | Crystal structure of FimH-LD R60P variant in the apo state | | Descriptor: | Protein FimH, SULFATE ION | | Authors: | Jakob, R.P, Rabbani, S, Ernst, B, Maier, T. | | Deposit date: | 2016-11-09 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Conformational switch of the bacterial adhesin FimH in the absence of the regulatory domain: Engineering a minimalistic allosteric system.
J. Biol. Chem., 293, 2018
|
|
4Z3G
 
 | | Crystal structure of the lectin domain of PapG from E. coli BI47 in complex with 4-methoxyphenyl beta-D-galabiose in space group P212121 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-methoxyphenol, PapG, ... | | Authors: | Jakob, R.P, Navarra, G, Zihlmann, P, Stangier, K, Preston, R.C, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2015-03-31 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Carbohydrate-Lectin Interactions: An Unexpected Contribution to Affinity.
Chembiochem, 18, 2017
|
|
4X5P
 
 | | Crystal structure of FimH in complex with a benzoyl-amidophenyl alpha-D-mannopyranoside | | Descriptor: | 4-{[3-chloro-4-(alpha-D-mannopyranosyloxy)phenyl]carbamoyl}benzoic acid, Protein FimH | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.997 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4X5Q
 
 | | Crystal structure of FimH in complex with 5-nitro-indolinylphenyl alpha-D-mannopyranoside | | Descriptor: | 4-(5-nitro-1H-indol-1-yl)phenyl alpha-D-mannopyranoside, Protein FimH | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4X50
 
 | | Crystal structure of FimH in complex with biphenyl alpha-D-mannopyranoside | | Descriptor: | Protein FimH, biphenyl-4-yl alpha-D-mannopyranoside | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-04 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4Z3H
 
 | | Crystal structure of the lectin domain of PapG from E. coli BI47 in complex with 4-methoxyphenyl beta-D-galabiose in space group P21 | | Descriptor: | 4-methoxyphenol, PapG, lectin domain, ... | | Authors: | Jakob, R.P, Navarra, G, Zihlmann, P, Stangier, K, Preston, R.C, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2015-03-31 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Carbohydrate-Lectin Interactions: An Unexpected Contribution to Affinity.
Chembiochem, 18, 2017
|
|
