6A5G
 
 | |
7VQ1
 
 | | Structure of Apo-hsTRPM2 channel | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
7VQ2
 
 | | Structure of Apo-hsTRPM2 channel TM domain | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
7WJ6
 
 | |
7WJ7
 
 | |
8GQU
 
 | | AK-42 inhibitor binding human ClC-2 TMD | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42.
Nat Commun, 14, 2023
|
|
7V9P
 
 | |
7V9Q
 
 | |
7V9O
 
 | |
7V9N
 
 | | Crystal structure of the lanthipeptide zinc-metallopeptidase EryP from saccharopolyspora erythraea in closed state | | Descriptor: | ACETATE ION, Alanine aminopeptidase, CALCIUM ION, ... | | Authors: | Zhao, C, Zhao, N.L, Bao, R. | | Deposit date: | 2021-08-26 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational remodeling enhances activity of lanthipeptide zinc-metallopeptidases.
Nat.Chem.Biol., 18, 2022
|
|
7EAX
 
 | | Crystal complex of p53-V272M and antimony ion | | Descriptor: | ANTIMONY (III) ION, Cellular tumor antigen p53, ZINC ION | | Authors: | Lu, M, Tang, Y. | | Deposit date: | 2021-03-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Repurposing antiparasitic antimonials to noncovalently rescue temperature-sensitive p53 mutations.
Cell Rep, 39, 2022
|
|
7DW5
 
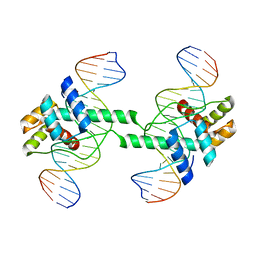 | | Crystal structure of DUX4 HD1-HD2 domain complexed with ERG sites | | Descriptor: | BROMIDE ION, DNA (5'-D(P*CP*GP*AP*CP*TP*TP*GP*AP*TP*GP*AP*GP*AP*TP*TP*AP*GP*AP*CP*TP*G)-3'), Double homeobox protein 4-like protein 2 | | Authors: | Zhang, H, Cheng, N, Li, Z, Zhang, W, Dong, X, Huang, J, Meng, G. | | Deposit date: | 2021-01-15 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | DNA crosslinking and recombination-activating genes 1/2 (RAG1/2) are required for oncogenic splicing in acute lymphoblastic leukemia.
Cancer Commun (Lond), 41, 2021
|
|
8HIV
 
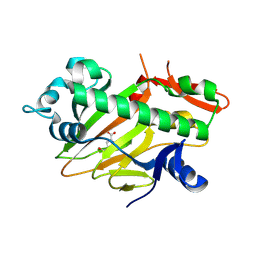 | | The structure of apo-SoBcmB with Fe(II) and AKG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, Fe/2OG dependent dioxygenase | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2022-11-22 | | Release date: | 2023-07-05 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2000308 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2U
 
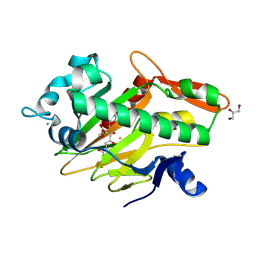 | | The complex structure of SoBcmB and its product 2f | | Descriptor: | (1S,5S,6S)-5-methyl-1-[(1S,2S)-2-methyl-1,2,3-tris(oxidanyl)propyl]-2-oxa-7,9-diazabicyclo[4.2.2]decane-8,10-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.00009918 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V34
 
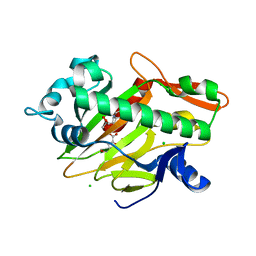 | | The complex structure of soBcmB and its product 1d | | Descriptor: | (3S,4S,5S,8S)-8-[(2S)-butan-2-yl]-3-methyl-3,4-bis(oxidanyl)-1-oxa-7,10-diazaspiro[4.5]decane-6,9-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.000169 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V3E
 
 | | The complex structure of soBcmB and its intermediate product 1a | | Descriptor: | (3S,6Z)-3-[(2S)-butan-2-yl]-6-[(2R)-2-methyl-2,3-bis(oxidanyl)propylidene]piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2T
 
 | | The complex structure of SoBcmB and its natural precursor 2 | | Descriptor: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Zhou, J.H, Wu, L. | | Deposit date: | 2021-08-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.20005679 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V36
 
 | | The complex structure of soBcmB and its intermediate product 2a | | Descriptor: | (3Z,6S)-3-[(2R)-2-methyl-2,3-bis(oxidanyl)propylidene]-6-[(2S)-4-oxidanylbutan-2-yl]piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.96009171 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V3N
 
 | | The complex structure of soBcmB-D307A and its natural precursor 2 | | Descriptor: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.850011 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2X
 
 | | The complex structure of soBcmB and its substrate 1 | | Descriptor: | (3S,6S)-3-[(2S)-butan-2-yl]-6-[(2R)-2-methyl-2,3-bis(oxidanyl)propyl]piperazine-2,5-dion, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.08387423 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V3O
 
 | | The structure of Se-SoBcmB with Fe(II)and AKG | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.83004665 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
8HMH
 
 | | The closed state of RGLG2-VWA | | Descriptor: | E3 ubiquitin-protein ligase RGLG2, MAGNESIUM ION | | Authors: | Wang, Q. | | Deposit date: | 2022-12-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The regulation of RGLG2-VWA by Ca 2+ ions.
Biochim Biophys Acta Proteins Proteom, 1872, 2024
|
|
8INH
 
 | | ZjOGT3, flavonoid 7,4'-di-O-glycosyltransferase | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, Z.L, Wang, H.D, Li, F.D, Ye, M. | | Deposit date: | 2023-03-09 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional characterization, structural basis, and regio-selectivity control of a promiscuous flavonoid 7,4'-di- O -glycosyltransferase from Ziziphus jujuba var. spinosa.
Chem Sci, 14, 2023
|
|
7D54
 
 | | Crstal structure MsGATase with Gln | | Descriptor: | GLUTAMINE, Glutamine amidotransferase class-I | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D4R
 
 | | SpuA native structure | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
