6TYW
 
 | |
6W8X
 
 | | Cryo-EM of the S. solfataricus pilus | | Descriptor: | pilin | | Authors: | Wang, F, Baquero, D.P, Su, Z, Beltran, L.C, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structures of two archaeal type IV pili illuminate evolutionary relationships.
Nat Commun, 11, 2020
|
|
6WIE
 
 | | Post-catalytic nicked complex of human Polymerase Mu on a complementary DNA double-strand break substrate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*G)-3'), ... | | Authors: | Kaminski, A.M, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2020-04-09 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of human DNA polymerase mu engaged on a DNA double-strand break.
Nat Commun, 11, 2020
|
|
4Q5N
 
 | |
4Q5F
 
 | |
4QGO
 
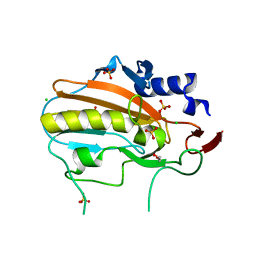 | | Crystal structure of NucA from Streptococcus agalactiae with no metal bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-entry nuclease (Competence-specific nuclease), ... | | Authors: | Pedersen, L.C, Moon, A.F, Gaudu, P. | | Deposit date: | 2014-05-23 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of the virulence factor nuclease A from Streptococcus agalactiae.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4R8H
 
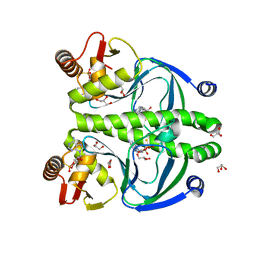 | | The role of protein-ligand contacts in allosteric regulation of the Escherichia coli Catabolite Activator Protein | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, GLYCEROL, cAMP-activated global transcriptional regulator CRP | | Authors: | Townsend, P.D, Pohl, E, McLeish, T.C.B, Rodgers, T.L, Glover, L.C, Korhonen, H.J, Wilson, M.R, Hodgson, D.R.W, Cann, M.J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The Role of Protein-Ligand Contacts in Allosteric Regulation of the Escherichia coli Catabolite Activator Protein.
J.Biol.Chem., 290, 2015
|
|
4U0F
 
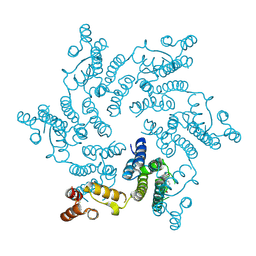 | | Hexameric HIV-1 CA in Complex with BI-2 | | Descriptor: | (4S)-4-(4-hydroxyphenyl)-3-phenyl-4,5-dihydropyrrolo[3,4-c]pyrazol-6(1H)-one, 1,2-ETHANEDIOL, Capsid protein p24 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
4U0B
 
 | |
4U0D
 
 | | Hexameric HIV-1 CA in complex with Nup153 peptide, P212121 crystal form | | Descriptor: | CHLORIDE ION, Gag polyprotein, Nuclear pore complex protein Nup153 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
4U0A
 
 | | Hexameric HIV-1 CA in complex with CPSF6 peptide, P6 crystal form | | Descriptor: | CHLORIDE ION, Capsid protein p24, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
4U0E
 
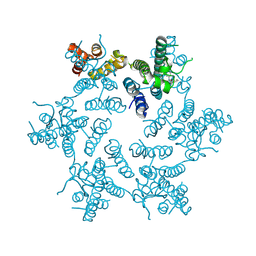 | | Hexameric HIV-1 CA in complex with PF3450074 | | Descriptor: | CHLORIDE ION, Capsid protein p24, N-METHYL-NALPHA-[(2-METHYL-1H-INDOL-3-YL)ACETYL]-N-PHENYL-L-PHENYLALANINAMIDE | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
4U0C
 
 | |
4UMC
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | L-PHOSPHOLACTATE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
4UCG
 
 | | NmeDAH7PS R126S variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE, ... | | Authors: | Cross, P.J, Heyes, L.C, Zhang, S, Nazmi, A.R, Parker, E.J. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Functional Unit of Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase is Dimeric.
Plos One, 11, 2016
|
|
4UC5
 
 | | Neisseria Meningitidis DAH7PS-Phenylalanine regulated | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, PHENYLALANINE, ... | | Authors: | Heyes, L.C, Lang, E.J.M, Parker, E.J. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Calculated Pka Variations Expose Dynamic Allosteric Communication Networks.
J.Am.Chem.Soc., 138, 2016
|
|
4UMB
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
4UMA
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3 deoxy D arabino heptulosonate 7 phosphate synthase the importance of accommodating the active site water | | Descriptor: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
7TLF
 
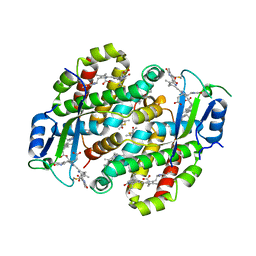 | | Structure of the photoacclimated Light Harvesting Complex PE545 from Proteomonas sulcata | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOERYTHROBILIN, Phycoerythrin alpha-subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7TJA
 
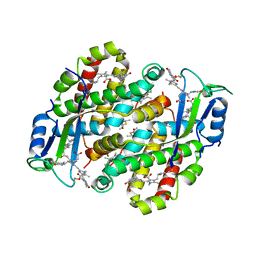 | | Structure of the Light Harvesting Complex PE545 from Proteomonas sulcata | | Descriptor: | 15,16-DIHYDROBILIVERDIN, MAGNESIUM ION, PHYCOERYTHROBILIN, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2022-01-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7T13
 
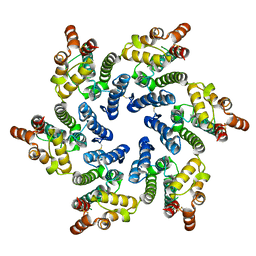 | |
7T12
 
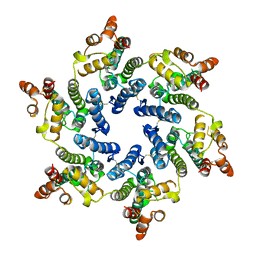 | | Hexameric HIV-1 (O-group) CA | | Descriptor: | Capsid protein p24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evasion of cGAS and TRIM5 defines pandemic HIV.
Nat Microbiol, 7, 2022
|
|
7T15
 
 | | Hexameric SIVcpz CA | | Descriptor: | Capsid protein p24 | | Authors: | Jacques, D.A, Dickson, C.F, James, L.C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Evasion of cGAS and TRIM5 defines pandemic HIV.
Nat Microbiol, 7, 2022
|
|
7T14
 
 | | Hexameric SIVmac CA | | Descriptor: | Capsid protein p24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evasion of cGAS and TRIM5 defines pandemic HIV.
Nat Microbiol, 7, 2022
|
|
8TCK
 
 | |
