5C2O
 
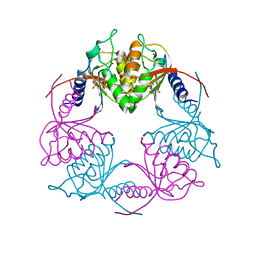 | | Crystal structure of Streptococcus mutans Deoxycytidylate Deaminase complexed with dTTP | | Descriptor: | MAGNESIUM ION, Putative deoxycytidylate deaminase, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Li, Y.H, Gao, Z.Q, Hou, H.F, Dong, Y.H. | | Deposit date: | 2015-06-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of the allosteric regulation of Streptococcus mutans 2'-deoxycytidylate deaminase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7F59
 
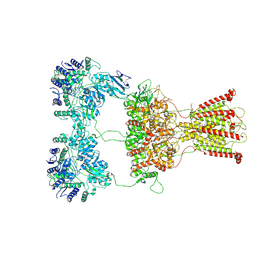 | | DNQX-bound GluK2-1xNeto2 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F56
 
 | | DNQX-bound GluK2-1xNeto2 complex, with asymmetric LBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F57
 
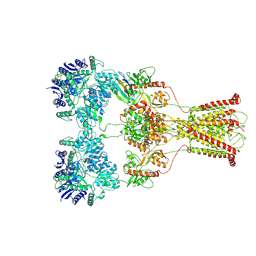 | | Kainate-bound GluK2-1xNeto2 complex, at the desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F5B
 
 | | LBD-TMD focused reconstruction of DNQX-bound GluK2-1xNeto2 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F5A
 
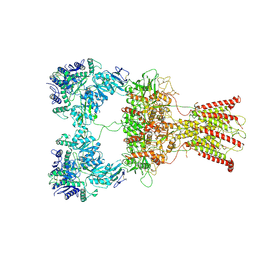 | | DNQX-bound GluK2-2xNeto2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
3KUY
 
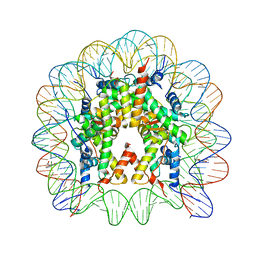 | |
7TDZ
 
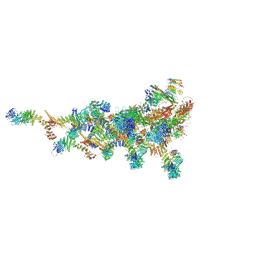 | |
7USG
 
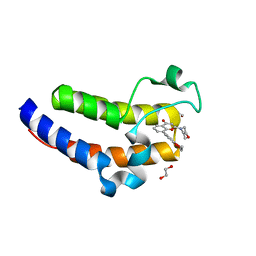 | | BRD2-BD2 in complex with MDP5 | | Descriptor: | (8M)-8-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-(morpholin-4-yl)-4H-1-benzopyran-4-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ... | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7USI
 
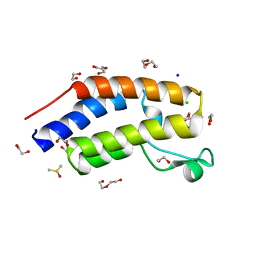 | | BRD2-BD1 in complex with MDP5 | | Descriptor: | (8M)-8-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-(morpholin-4-yl)-4H-1-benzopyran-4-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ... | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7USH
 
 | | BRD2-BD2 in complex with SF2523 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 2 | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7USK
 
 | | BRD4-BD2 Ligand free | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7USJ
 
 | | BRD4-BD2 in complex with SF2523 | | Descriptor: | 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 4 | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
2EXX
 
 | | Crystal structure of HSCARG from Homo sapiens in complex with NADP | | Descriptor: | GLYCEROL, HSCARG protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dai, X, Chen, Q, Yao, D, Liang, Y, Dong, Y, Gu, X, Zheng, X, Luo, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Restructuring of the dinucleotide-binding fold in an NADP(H) sensor protein.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2NB4
 
 | | Solution structure of Q388A3 PDZ domain | | Descriptor: | Putative uncharacterized protein | | Authors: | Mei, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Q388A3 PDZ domain from Trypanosoma brucei
J.Struct.Biol., 194, 2016
|
|
5XOB
 
 | | Crystal structure of apo TiaS (tRNAIle2 agmatidine synthetase) | | Descriptor: | MAGNESIUM ION, ZINC ION, tRNA(Ile2) 2-agmatinylcytidine synthetase TiaS | | Authors: | Dong, J. | | Deposit date: | 2017-05-27 | | Release date: | 2018-08-29 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of tRNA-Modifying Enzyme TiaS and Motions of Its Substrate Binding Zinc Ribbon.
J. Mol. Biol., 430, 2018
|
|
4JKZ
 
 | | Crystal structure of ms6564 from mycobacterium smegmatis | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Yang, S.F, Gao, Z.Q, He, Z.G, Dong, Y.H. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for interaction between Mycobacterium smegmatis Ms6564, a TetR family master regulator, and its target DNA.
J.Biol.Chem., 288, 2013
|
|
4JL3
 
 | | Crystal structure of ms6564-dna complex | | Descriptor: | DNA (31-MER), Transcriptional regulator, TetR family | | Authors: | Yang, S.F, Gao, Z.Q, He, Z.G, Dong, Y.H. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-26 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for interaction between Mycobacterium smegmatis Ms6564, a TetR family master regulator, and its target DNA.
J.Biol.Chem., 288, 2013
|
|
7DYM
 
 | | Pseudomonas aeruginosa TseT-TsiT complex | | Descriptor: | Imm52 domain-containing protein, Tox-REase-5 domain-containing protein | | Authors: | She, Z. | | Deposit date: | 2021-01-22 | | Release date: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and SAXS studies unveiled a novel inhibition mechanism of the Pseudomonas aeruginosa T6SS TseT-TsiT complex.
Int.J.Biol.Macromol., 188, 2021
|
|
7FFT
 
 | |
7FFW
 
 | | The crystal structure of a domain-swapped dimeric maltodextrin-binding protein MalE from Salmonella enterica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, METHOXYETHANE, ... | | Authors: | Wang, L, Bu, T, Bai, X. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the domain-swapped dimeric maltodextrin-binding protein MalE from Salmonella enterica.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4NG2
 
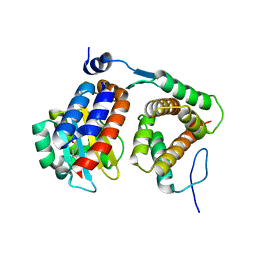 | |
4O7P
 
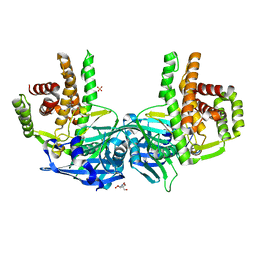 | | Crystal structure of Mycobacterium tuberculosis maltose kinase MaK complexed with maltose | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Maltokinase, SULFATE ION, ... | | Authors: | Li, J, Guan, X.T, Rao, Z.H. | | Deposit date: | 2013-12-26 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Homotypic dimerization of a maltose kinase for molecular scaffolding.
Sci Rep, 4, 2014
|
|
4O7O
 
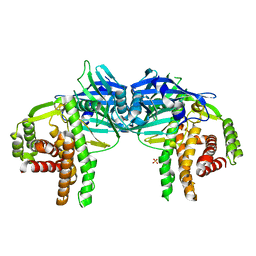 | |
5YS1
 
 | | Crystal structure of Multicopper Oxidase CueO G304K mutant | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Wang, H.Q, Liu, X.Q, Zhao, J.T, Yue, Q.X, Yan, Y.H, Dong, Y.H, Fan, Y.L, Tian, J, Wu, N.F, Gong, Y. | | Deposit date: | 2017-11-12 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structures of multicopper oxidase CueO G304K mutant: structural basis of the increased laccase activity
Sci Rep, 8, 2018
|
|
