2I83
 
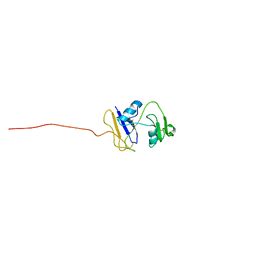 | | hyaluronan-binding domain of CD44 in its ligand-bound form | | Descriptor: | CD44 antigen | | Authors: | Takeda, M, Ogino, S, Umemoto, R, Sakakura, M, Kajiwara, M, Sugahara, K.N, Hayasaka, H, Miyasaka, M, Terasawa, H, Shimada, I. | | Deposit date: | 2006-09-01 | | Release date: | 2006-11-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Ligand-induced Structural Changes of the CD44 Hyaluronan-binding Domain Revealed by NMR
J.Biol.Chem., 281, 2006
|
|
1MRB
 
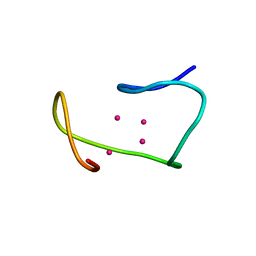 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
5LQF
 
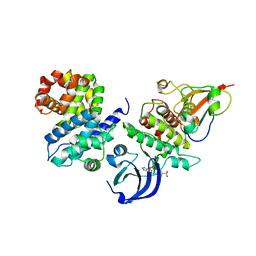 | | CDK1/CyclinB1/CKS2 in complex with NU6102 | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B.J, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E, Newell, D.R, Turner, D.M, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Endicott, J.A, Cano, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
7WAF
 
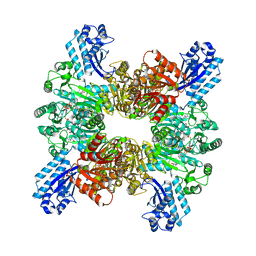 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS and 4x(beta-Asp-Arg) | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, Cyanophycin synthase, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7WAE
 
 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS, 4x(beta-Asp-Arg), and aspartate | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, ASPARTIC ACID, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
5NEV
 
 | | CDK2/Cyclin A in complex with compound 73 | | Descriptor: | 4-[[6-(3-phenylphenyl)-7~{H}-purin-2-yl]amino]benzenesulfonamide, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E.M, Newell, D.R, Turner, D, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Cano, G. | | Deposit date: | 2017-03-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | Descriptor: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.61 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
7WLG
 
 | | Cryo-EM structure of GH31 alpha-1,3-glucosidase from Lactococcus lactis subsp. cremoris | | Descriptor: | Alpha-xylosidase | | Authors: | Ikegaya, M, Moriya, T, Adachi, N, Kawasaki, M, Park, E.Y, Miyazaki, T. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
4LW9
 
 | | Crystal structure of Vibrio cholera major pseudopilin EpsG | | Descriptor: | CALCIUM ION, CHLORIDE ION, PLATINUM (II) ION, ... | | Authors: | Vago, F.S, Raghunathan, K, Jens, J.C, Wedemeyer, W.J, Bagdasarian, M, Brunzelle, J.S, Arvidson, D.N. | | Deposit date: | 2013-07-26 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Vibrio cholera major pseudopilin EpsG
To be Published
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.91 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
6JNA
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JN9
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JNC
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JND
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
5DE0
 
 | | Dye-decolorizing protein from V. cholerae | | Descriptor: | Deferrochelatase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Uchida, T, Sasaki, M, Tanaka, Y, Yao, M. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A Dye-Decolorizing Peroxidase from Vibrio cholerae.
Biochemistry, 54, 2015
|
|
2MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD-7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
5B3I
 
 | | Homo-dimeric structure of cytochrome c' from Thermophilic Hydrogenophilus thermoluteolus | | Descriptor: | Cytochrome c prime, HEME C | | Authors: | Fujii, S, Oki, H, Kawahara, K, Yamane, D, Yamanaka, M, Maruno, T, Kobayashi, Y, Masanari, M, Wakai, S, Nishihara, H, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and functional insights into thermally stable cytochrome c' from a thermophile
Protein Sci., 26, 2017
|
|
6T0W
 
 | | Human Influenza B polymerase recycling complex | | Descriptor: | Polymerase PB2, Polymerase acidic protein, RNA-directed RNA polymerase catalytic subunit, ... | | Authors: | Wandzik, J.M, Kouba, T, Karuppasamy, M, Cusack, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
