2GRB
 
 | |
8E14
 
 | | Cryo-EM structure of Rous sarcoma virus strand transfer complex | | Descriptor: | DNA (42-MER), DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*TP*CP*TP*TP*CP*TP*TP*TP*C)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular determinants for Rous sarcoma virus intasome assemblies involved in retroviral integration.
J.Biol.Chem., 299, 2023
|
|
8E5D
 
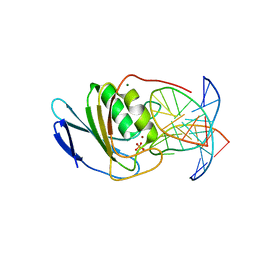 | | Crystal structure of double-stranded DNA deaminase toxin DddA in complex with DNA with the target cytosine parked in the major groove | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*CP*GP*GP*AP*CP*GP*TP*TP*GP*C)-3'), Double-stranded DNA deaminase toxin A, MAGNESIUM ION, ... | | Authors: | Yin, L.L, Shi, K, Aihara, H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural basis of sequence-specific cytosine deamination by double-stranded DNA deaminase toxin DddA.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8E5E
 
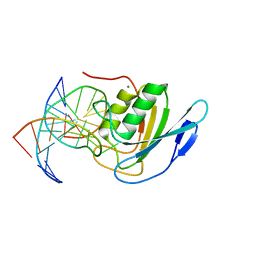 | | Crystal structure of double-stranded DNA deaminase toxin DddA in complex with DNA with the target cytosine flipped into the active site | | Descriptor: | DNA (5'-D(*GP*CP*AP*AP*CP*GP*TP*CP*CP*GP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*CP*GP*GP*AP*CP*GP*TP*TP*GP*C)-3'), Double-stranded DNA deaminase toxin A, ... | | Authors: | Yin, L.L, Shi, K, Aihara, H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis of sequence-specific cytosine deamination by double-stranded DNA deaminase toxin DddA.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EUO
 
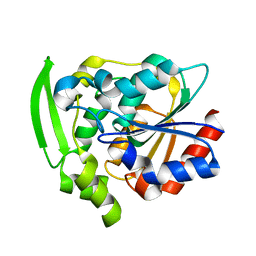 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seven Mutations | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Greenberg, L.R, Walsh, M.E, Kazlauskas, R.J, Pierce, C.T, Shi, K, Aihara, H, Evans, R.L. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | to be published
To Be Published
|
|
8EJP
 
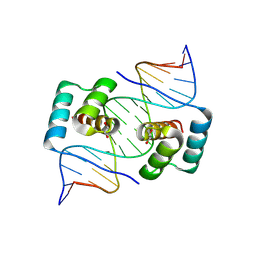 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA containing 5-Bromouracil | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Yin, L.L, Shi, K, Aihara, H. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8EJO
 
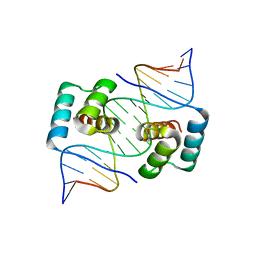 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Yin, L.L, Shi, K, Aihara, H. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8FR5
 
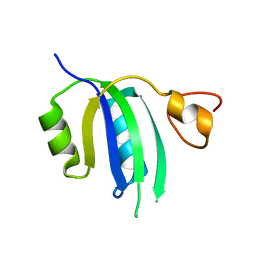 | | Crystal structure of the Human Smacovirus 1 Rep domain | | Descriptor: | MANGANESE (II) ION, Rep, SODIUM ION | | Authors: | Limon, L.K, Shi, K, Dao, A, Rugloski, J, Tompkins, K.J, Aihara, H, Gordon, W.R, Evans IIII, R.L. | | Deposit date: | 2023-01-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The crystal structure of the human smacovirus 1 Rep domain.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
5IRS
 
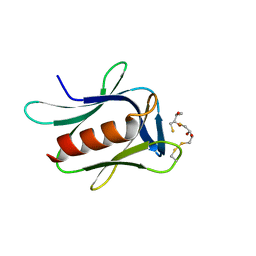 | | crystal structure of the proteasomal Rpn13 PRU-domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Proteasomal ubiquitin receptor ADRM1 | | Authors: | Chen, X, Shi, K, Walters, K, Aihara, H. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
1J6S
 
 | | Crystal Structure of an RNA Tetraplex (UGAGGU)4 with A-tetrads, G-tetrads, U-tetrads and G-U octads | | Descriptor: | 5'-R(*(BRUP*GP*AP*GP*GP*U)-3', BARIUM ION, SODIUM ION | | Authors: | Pan, B, Xiong, Y, Shi, K, Deng, J, Sundaralingam, M. | | Deposit date: | 2002-07-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an RNA purine-rich tetraplex containing adenine tetrads:
implications for specific binding in RNA tetraplexes
Structure, 11, 2003
|
|
4EP4
 
 | | Thermus thermophilus RuvC structure | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, GLYCEROL, MAGNESIUM ION | | Authors: | Chen, L, Shi, K, Yin, Z.Q, Aihara, H. | | Deposit date: | 2012-04-17 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution.
Nucleic Acids Res., 41, 2013
|
|
1IK5
 
 | | Crystal Structure of a 14mer RNA Containing Double UU Bulges in Two Crystal Forms: A Novel U*(AU) Intramolecular Base Triple | | Descriptor: | 5'-R(*GP*GP*UP*AP*UP*UP*UP*CP*GP*GP*UP*AP*(CBR)P*C)-3', 5'-R(*GP*GP*UP*AP*UP*UP*UP*UP*GP*GP*UP*AP*(CBR)P*C)-3', MAGNESIUM ION | | Authors: | Deng, J, Xiong, Y, Sudarsanakumar, C, Shi, K, Sundaralingam, M. | | Deposit date: | 2001-05-02 | | Release date: | 2001-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two forms of a 14-mer RNA/DNA chimer duplex with double UU bulges: a novel intramolecular U*(A x U) base triple.
RNA, 7, 2001
|
|
4EP5
 
 | | Thermus thermophilus RuvC structure | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, GLYCEROL, SULFATE ION | | Authors: | Chen, L, Shi, K, Yin, Z.Q, Aihara, H. | | Deposit date: | 2012-04-17 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution.
Nucleic Acids Res., 41, 2013
|
|
1MDG
 
 | | An Alternating Antiparallel Octaplex in an RNA Crystal Structure | | Descriptor: | 5'-R(*UP*(BGM)GP*AP*GP*GP*U)-3', COBALT HEXAMMINE(III), SODIUM ION | | Authors: | Pan, B.C, Xiong, Y, Shi, K, Sundaralingam, M. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Eight-Stranded Helical Fragment in RNA Crystal Structure: Implications for Tetraplex Interaction
Structure, 11, 2003
|
|
5WLY
 
 | | E. coli LpxH- 8 mutations | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The substrate-binding cap of the UDP-diacylglucosamine pyrophosphatase LpxH is highly flexible, enabling facile substrate binding and product release.
J. Biol. Chem., 293, 2018
|
|
7M8R
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with fluorosubstituted tryptophans | | Descriptor: | 1,1,1-tris(fluoranyl)propan-2-one, 1,2-ETHANEDIOL, BENZOIC ACID, ... | | Authors: | Johns, J.C, Banerjee, R, Shi, K, Semonis, M.M, Aihara, H, Pomerantz, W.C.K, Lipscomb, J.D. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Soluble Methane Monooxygenase Component Interactions Monitored by 19 F NMR.
Biochemistry, 60, 2021
|
|
7M8Q
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with fluorosubstituted tryptophans | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Shi, K, Semonis, M.M, Aihara, H, Pomerantz, W.C.K, Lipscomb, J.D. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Soluble Methane Monooxygenase Component Interactions Monitored by 19 F NMR.
Biochemistry, 60, 2021
|
|
7MLR
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(4-{5-[6-(3,5-dimethylphenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1- yl}piperidin-1-yl)-N,N-dimethylethan-1-amine (DW34) | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-{5-[6-(3,5-dimethylphenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine, Bromodomain-containing protein 4, ... | | Authors: | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
7MLS
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine, Bromodomain-containing protein 4, ... | | Authors: | Cui, H, Johnson, J.A, Zahid, H, Buchholz, C.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
7MLQ
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine (compound 26) | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine, Bromodomain-containing protein 4, ... | | Authors: | Cui, H, Johnson, J.A, Vail, N.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
3HL6
 
 | | Staphylococcus aureus pathogenicity island 3 ORF9 protein | | Descriptor: | CHLORIDE ION, Pathogenicity island protein | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Schlievert, P.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional studies of a pathogenicity island protein from Staphylococcus aureus
To be Published
|
|
3K55
 
 | | Structure of beta hairpin deletion mutant of beta toxin from Staphylococcus aureus | | Descriptor: | Beta-hemolysin, CHLORIDE ION, SODIUM ION | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-10-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure of a mutant beta toxin from Staphylococcus aureus reveals domain swapping and conformational flexibility
Acta Crystallogr.,Sect.F, 67, 2011
|
|
418D
 
 | |
2AWE
 
 | |
4NQ3
 
 | | Crystal structure of cyanuic acid hydrolase from A. caulinodans | | Descriptor: | BARBITURIC ACID, Cyanuric acid amidohydrolase, MAGNESIUM ION, ... | | Authors: | Cho, S, Shi, K, Aihara, H. | | Deposit date: | 2013-11-23 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Cyanuric acid hydrolase from Azorhizobium caulinodans ORS 571: crystal structure and insights into a new class of Ser-Lys dyad proteins.
Plos One, 9, 2014
|
|
