8FRQ
 
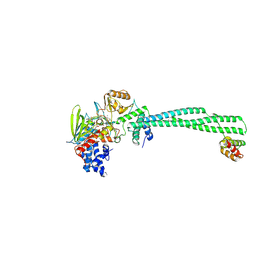 | | LSD1-CoREST in complex with T14, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(4aS)-7,8-dimethyl-5-(3-{4-[(5-methyl-1,3,4-thiadiazol-2-yl)carbamoyl]phenyl}propanoyl)-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-01-08 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
8FRV
 
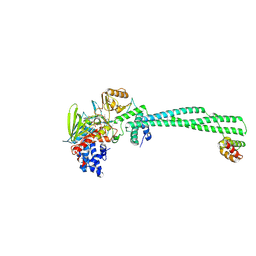 | | LSD1-CoREST in complex with T17, short soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,13R)-3-[3-(dimethylcarbamoyl)phenyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-01-09 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To be published
|
|
8FRI
 
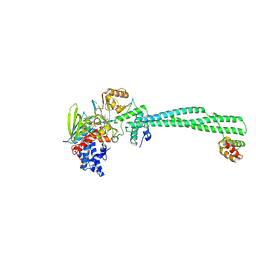 | | LSD1-CoREST in complex with AW4, short soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3S,3aS,13R)-1-hydroxy-10,11-dimethyl-4,6-dioxo-3-([1~1~,2~1~:2~3~,3~1~-terphenyl]-1~4~-yl)-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-01-07 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8FSK
 
 | | LSD1-CoREST in complex with T18, short soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,13R)-3-(3-benzamidophenyl)-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-01-10 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
1KNR
 
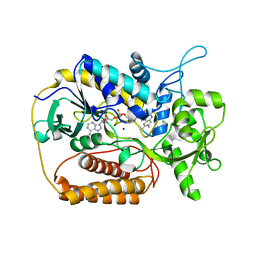 | | L-aspartate oxidase: R386L mutant | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, L-aspartate oxidase, ... | | Authors: | Bossi, R.T, Mattevi, A. | | Deposit date: | 2001-12-19 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of FAD-bound L-aspartate oxidase: insight into substrate specificity and catalysis.
Biochemistry, 41, 2002
|
|
1KNP
 
 | |
6ZL6
 
 | |
6ZLD
 
 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Glucuronic acid and NAD | | Descriptor: | Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Iacovino, L.G, Savino, S, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
6ZLL
 
 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Galacturonic acid and NAD | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Iacovino, L.G, Savino, S, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
7AL4
 
 | | Ancestral Flavin-containing monooxygenase (FMO) 1 (mammalian) | | Descriptor: | Ancestral Flavin-containing monooxygenase 1 (mammalian), CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nicoll, C.R, Mattevi, A. | | Deposit date: | 2020-10-05 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ancestral reconstruction of mammalian FMO1 enables structural determination, revealing unique features that explain its catalytic properties.
J.Biol.Chem., 296, 2020
|
|
2C7G
 
 | | FprA from Mycobacterium tuberculosis: His57Gln mutant | | Descriptor: | 4-OXO-NICOTINAMIDE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-FERREDOXIN REDUCTASE FPRA, ... | | Authors: | Pennati, A, Razeto, A, De Rosa, M, Pandini, V, Vanoni, M.A, Aliverti, A, Mattevi, A, Coda, A, Zanetti, G. | | Deposit date: | 2005-11-24 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the His57-Glu214 Ionic Couple Located in the Active Site of Mycobacterium Tuberculosis Fpra.
Biochemistry, 45, 2006
|
|
6ZLA
 
 | |
6ZLK
 
 | | Equilibrium Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Glucuronic acid/UDP-Galacturonic acid and NAD | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
6SZ5
 
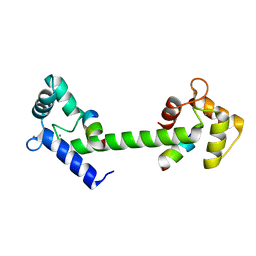 | |
1A70
 
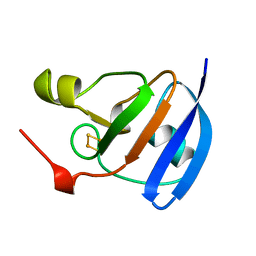 | | SPINACH FERREDOXIN | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Binda, C, Coda, A, Mattevi, A, Aliverti, A, Zanetti, G. | | Deposit date: | 1998-03-19 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the mutant E92K of [2Fe-2S] ferredoxin I from Spinacia oleracea at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BXS
 
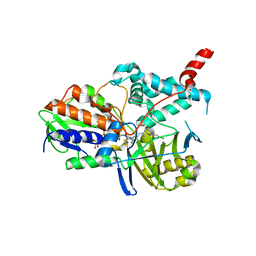 | | Human Monoamine Oxidase A in complex with Clorgyline, Crystal Form B | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] A, FLAVIN-ADENINE DINUCLEOTIDE, N-[3-(2,4-DICHLOROPHENOXY)PROPYL]-N-METHYL-N-PROP-2-YNYLAMINE | | Authors: | De Colibus, L, Binda, C, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2005-07-27 | | Release date: | 2005-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Three-Dimensional Structure of Human Monoamine Oxidase a (Mao A): Relation to the Structures of Rat Mao a and Human Mao B
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BYB
 
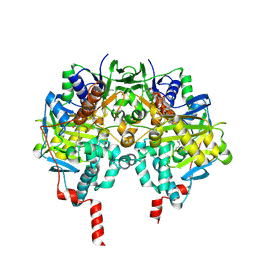 | | Human Monoamine Oxidase B in complex with Deprenyl | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] B, DEPRENYL, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, De Colibus, L, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2005-07-29 | | Release date: | 2005-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-Dimensional Structure of Human Monoamine Oxidase a (Mao A): Relation to the Structures of Rat Mao a and Human Mao B
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4B68
 
 | | A. fumigatus ornithine hydroxylase (SidA), re-oxidised state bound to NADP and Arg | | Descriptor: | ARGININE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Franceschini, S, Fedkenheuer, M, Vogelaar, N.J, Robinson, H.H, Sobrado, P, Mattevi, A. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Insight Into the Mechanism of Oxygen Activation and Substrate Selectivity of Flavin-Dependent N-Hydroxylating Monooxygenases.
Biochemistry, 51, 2012
|
|
6H0N
 
 | |
6HHE
 
 | | Crystal structure of the medfly Odorant Binding Protein CcapOBP22/CcapOBP69a | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Odorant binding protein OBP69a, SULFATE ION | | Authors: | Falchetto, M, Ciossani, G, Nenci, S, Mattevi, A, Gasperi, G, Forneris, F. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.516 Å) | | Cite: | Structural and biochemical evaluation of Ceratitis capitata odorant-binding protein 22 affinity for odorants involved in intersex communication.
Insect Mol.Biol., 28, 2019
|
|
6HQD
 
 | | Cytochrome P450-153 from Pseudomonas sp. 19-rlim | | Descriptor: | Cytochrome P450, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-24 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
6HQW
 
 | |
6H0P
 
 | |
1B5Q
 
 | | A 30 ANGSTROM U-SHAPED CATALYTIC TUNNEL IN THE CRYSTAL STRUCTURE OF POLYAMINE OXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, N,N'-BIS(2,3-BUTADIENYL)-1,4-BUTANE-DIAMINE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase.
Structure Fold.Des., 7, 1999
|
|
1B37
 
 | | A 30 ANGSTROM U-SHAPED CATALYTIC TUNNEL IN THE CRYSTAL STRUCTURE OF POLYAMINE OXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (POLYAMINE OXIDASE), ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 1998-12-17 | | Release date: | 1999-12-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase.
Structure Fold.Des., 7, 1999
|
|
