1KK9
 
 | | CRYSTAL STRUCTURE OF E. COLI YCIO | | Descriptor: | SULFATE ION, probable translation factor yciO | | Authors: | Jia, J, Lunin, V.V, Sauve, V, Huang, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-12-06 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the YciO protein from Escherichia coli
PROTEINS: STRUCT.,FUNCT.,GENET., 49, 2002
|
|
7UCS
 
 | |
7UCT
 
 | |
7UD3
 
 | |
7UCV
 
 | |
7UCZ
 
 | |
7UD1
 
 | |
6H38
 
 | | The crystal structure of human carbonic anhydrase VII in complex with 4-[(4-fluorophenyl)methyl]-1-piperazinyl]benzenesulfonamide. | | Descriptor: | 4-[4-[(4-fluorophenyl)methyl]piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 7, GLYCEROL, ... | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
7UCN
 
 | |
1HLK
 
 | | METALLO-BETA-LACTAMASE FROM BACTEROIDES FRAGILIS IN COMPLEX WITH A TRICYCLIC INHIBITOR | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, BETA-LACTAMASE, TYPE II, ... | | Authors: | Payne, D.J, Hueso-Rodriguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Chever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Diez, E, Perez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2000-12-01 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad spectrum inhibitors of metallo-beta-lactamases
Antimicrob.Agents Chemother., 46, 2002
|
|
3DT5
 
 | | C_terminal domain of protein of unknown function AF_0924 from Archaeoglobus fulgidus. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Uncharacterized protein AF_0924 | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-14 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray crystal structure of C_terminal domain of protein of unknown function AF_0924 from Archaeoglobus fulgidus.
To be Published
|
|
6H36
 
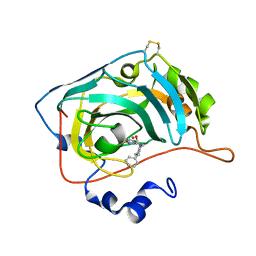 | | The crystal structure of human carbonic anhydrase VII in complex with 4-(4-phenylpiperidine-1-carbonyl)benzenesulfonamide. | | Descriptor: | 4-(4-phenylpiperidin-1-yl)carbonylbenzenesulfonamide, Carbonic anhydrase 7, ZINC ION | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
1HLY
 
 | | SOLUTION STRUCTURE OF HONGOTOXIN 1 | | Descriptor: | HONGOTOXIN 1 | | Authors: | Pragl, B, Koschak, A, Trieb, M, Obermair, G, Kaufmann, W.A, Gerster, U, Blanc, E, Hahn, C, Prinz, H, Schutz, G, Darbon, H, Gruber, H.J, Knaus, H.G. | | Deposit date: | 2000-12-04 | | Release date: | 2003-09-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Synthesis, characterization, and application of cy-dye- and alexa-dye-labeled hongotoxin(1) analogues. The
first high affinity fluorescence probes for voltage-gated K+ channels.
BIOCONJUG.CHEM., 13, 2002
|
|
1ZVL
 
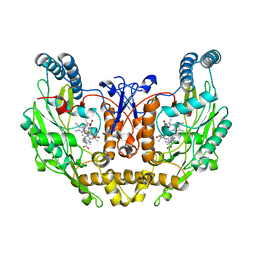 | | Rat Neuronal Nitric Oxide Synthase Oxygenase Domain complexed with natural substrate L-Arg. | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, Nitric-oxide synthase, ... | | Authors: | Matter, H, Kumar, H.S, Fedorov, R, Frey, A, Kotsonis, P, Hartmann, E, Frohlich, L.G, Reif, A, Pfleiderer, W, Scheurer, P, Ghosh, D.K, Schlichting, I, Schmidt, H.H. | | Deposit date: | 2005-06-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of Isoform-Specific Inhibitors Targeting the Tetrahydrobiopterin Binding Site of Human Nitric Oxide Synthases.
J.Med.Chem., 48, 2005
|
|
2PC6
 
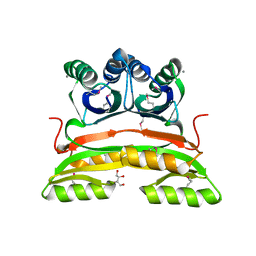 | | Crystal structure of putative acetolactate synthase- small subunit from Nitrosomonas europaea | | Descriptor: | CALCIUM ION, Probable acetolactate synthase isozyme III (Small subunit), UNKNOWN LIGAND | | Authors: | Petkowski, J.J, Chruszcz, M, Zimmerman, M.D, Zheng, H, Cymborowski, M.T, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of TM0549 and NE1324--two orthologs of E. coli AHAS isozyme III small regulatory subunit.
Protein Sci., 16, 2007
|
|
3E1L
 
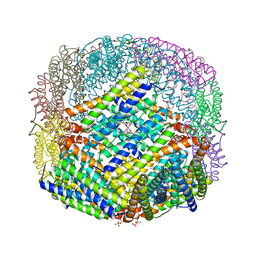 | | Crystal structure of E. coli Bacterioferritin (BFR) soaked in phosphate with an alternative conformation of the unoccupied Ferroxidase centre (APO-BFR II). | | Descriptor: | BACTERIOFERRITIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Crow, A, Lawson, T, Lewin, A, Moore, G.R, Le Brun, N. | | Deposit date: | 2008-08-04 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for iron mineralization by bacterioferritin
J.Am.Chem.Soc., 131, 2009
|
|
2AEY
 
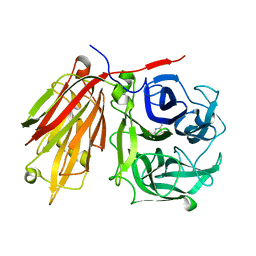 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus in complex with 2,5 dideoxy-2,5-immino-D-mannitol | | Descriptor: | 2,5-DIDEOXY-2,5-IMINO-D-MANNITOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Verhaest, M, Le Roy, K, De Ranter, C.J, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor.
New Phytol, 174, 2007
|
|
2W9O
 
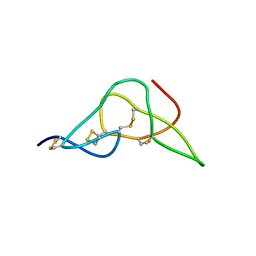 | | Solution structure of jerdostatin from Trimeresurus jerdonii | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-27 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9U
 
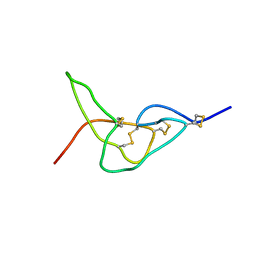 | | Solution structure of jerdostatin mutant R24K from Trimeresurus jerdonii | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9V
 
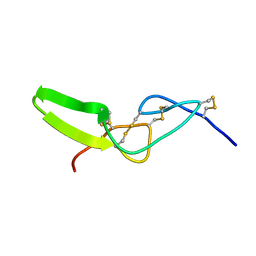 | | Solution structure of jerdostatin from Trimeresurus jerdonii with end C-terminal residues N45G46 deleted | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9W
 
 | | Solution structure of jerdostatin mutant R24K from Trimeresurus jerdonii with end C-terminal residues N45G46 deleted | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
1HWV
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-10 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
1HX4
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
3H0K
 
 | |
2ZGA
 
 | | HIV-1 protease in complex with a dimethylallyl decorated pyrrolidine based inhibitor (hexagonal space group) | | Descriptor: | (3S,4S),-3,4-Bis-[(4-carbamoyl-benzensulfonyl)-(3-methyl-but-2-enyl)-amino]-pyrrolidine, Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2008-01-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Solutions for the Same Problem: Multiple Binding Modes of Pyrrolidine-Based HIV-1 Protease Inhibitors
J.Mol.Biol., 410, 2011
|
|
