5DDG
 
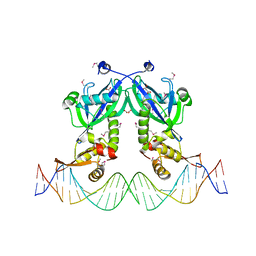 | | The structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with target double strand DNA | | Descriptor: | DNA (27-MER), FORMIC ACID, MALONIC ACID, ... | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
4EYG
 
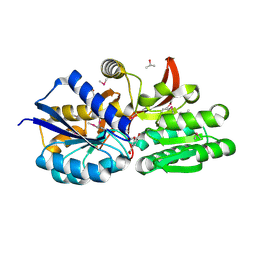 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris BisB5 in complex with vanillic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXY-3-METHOXYBENZOATE, ISOPROPYL ALCOHOL, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
4ER9
 
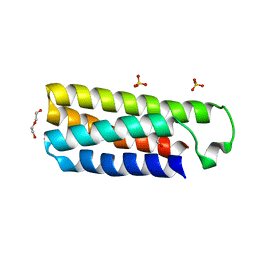 | | Crystal structure of cytochrome b562 from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Jedrzejczak, R, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Crystal structure of cytochrome b562 from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S
To be Published
|
|
4E4R
 
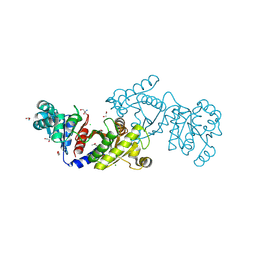 | | EutD phosphotransacetylase from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | EutD phosphotransacetylase from Staphylococcus aureus.
To be Published
|
|
5DN1
 
 | | Crystal structure of Phosphoribosyl isomerase A from Streptomyces coelicolor | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, GLYCEROL, Phosphoribosyl isomerase A, ... | | Authors: | Chang, C, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-09 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
4EH1
 
 | | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoprotein, ... | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
5EWQ
 
 | | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames | | Descriptor: | ACETATE ION, Amidase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames
To Be Published
|
|
5F1P
 
 | | Crystal Structure of Dehydrogenase from Streptomyces platensis | | Descriptor: | PtmO8 | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.-Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-11-30 | | Release date: | 2015-12-30 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal Structure of a Dehydrogenase, PtmO8, from Streptomyces platensis
To Be Published
|
|
5EV7
 
 | | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames | | Descriptor: | Conserved domain protein | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames.
To Be Published
|
|
4GYQ
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase NDM-1, MAGNESIUM ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae
To be Published, 2012
|
|
5EYF
 
 | | Crystal Structure of Solute-binding Protein from Enterococcus faecium with Bound Glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, Glutamate ABC superfamily ATP binding cassette transporter, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-24 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Solute-binding Protein from Enterococcus faecium with Bound Glutamate
To Be Published
|
|
4H3V
 
 | | Crystal structure of oxidoreductase domain protein from Kribbella flavida | | Descriptor: | FORMIC ACID, Oxidoreductase domain protein | | Authors: | Michalska, K, Mack, J.C, McKnight, S.M, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-14 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of oxidoreductase domain protein from Kribbella flavida
To be Published
|
|
4HAO
 
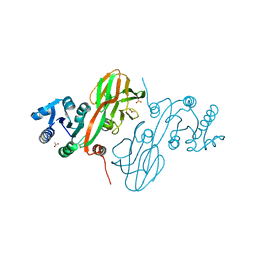 | | Crystal Structure of Inorganic Polyphosphate/ATP-NAD Kinase from Yersinia pestis CO92 | | Descriptor: | ACETIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Crystal Structure of Inorganic Polyphosphate/ATP-NAD Kinase from Yersinia pestis CO92
To be Published
|
|
4HG2
 
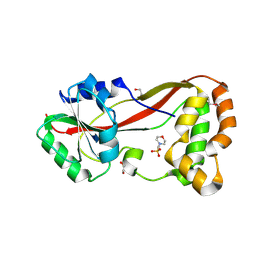 | | The Structure of a Putative Type II Methyltransferase from Anaeromyxobacter dehalogenans. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Methyltransferase type 11 | | Authors: | Cuff, M.E, Chhor, G, Clancy, S, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-10-05 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of a Putative Type II Methyltransferase from Anaeromyxobacter dehalogenans.
TO BE PUBLISHED
|
|
5F1Y
 
 | |
5F6Q
 
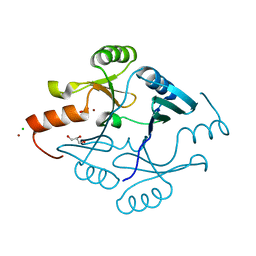 | | Crystal Structure of Metallothiol Transferase from Bacillus anthracis str. Ames | | Descriptor: | CHLORIDE ION, GLYCEROL, Metallothiol transferase FosB 2, ... | | Authors: | Maltseva, N, Kim, Y, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-06 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Metallothiol Transferase from Bacillus anthracis str. Ames
To Be Published
|
|
5F3K
 
 | | X-Ray Crystallographic Structure of hTrap1 N-terminal Domain-apo | | Descriptor: | Heat shock protein 75 kDa, mitochondrial | | Authors: | Sung, N, Lee, J, Kim, J, Chang, C, Joachimiak, A, Lee, S, Tsai, F.T.F. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4GPN
 
 | | The crystal structure of 6-P-beta-D-Glucosidase (E375Q mutant) from Streptococcus mutans UA150 in complex with Gentiobiose 6-phosphate. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose-(1-6)-beta-D-glucopyranose, 6-phospho-beta-D-Glucosidase, ... | | Authors: | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4GQT
 
 | | N-terminal domain of C. elegans Hsp90 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 90, ZINC ION | | Authors: | Osipiuk, J, Chhor, G, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-23 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | N-terminal domain of C. elegans Hsp90
To be Published
|
|
2I17
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594 at temperature of 60K | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Mitshler, A, Hasemann, I, Schneider, T, Cousido, A, Lunin, V.Y, Joachimiak, A, Podjarny, A. | | Deposit date: | 2006-08-13 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Ultrahigh-resolution study of protein atomic displacement parameters at cryotemperatures obtained with a helium cryostat.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4GZE
 
 | | Crystal structure of 6-phospho-beta-glucosidase from Lactobacillus plantarum (apo form) | | Descriptor: | 6-phospho-beta-glucosidase, CHLORIDE ION, GLYCEROL | | Authors: | Michalska, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-06 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HB7
 
 | | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50. | | Descriptor: | 1,2-ETHANEDIOL, Dihydropteroate synthase | | Authors: | Cuff, M.E, Holowicki, J, Jedrzejczak, R, Terwilliger, T.C, Rubin, E.J, Guinn, K, Baker, D, Ioerger, T.R, Sacchettini, J.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50.
TO BE PUBLISHED
|
|
3D1R
 
 | | Structure of E. coli GlpX with its substrate fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Singer, A, Skarina, T, Dong, A, Brown, G, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
5FAR
 
 | | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complex with 9-METHYLGUANINE | | Descriptor: | 7,8-dihydroneopterin aldolase, 9-METHYLGUANINE | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complex with 9-METHYLGUANINE
To Be Published
|
|
2I16
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594 at temperature of 15K | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Petrova, T, Ginell, S, Mitshler, A, Hasemann, I, Schneider, T, Cousido, A, Lunin, V.Y, Joachimiak, A, Podjarny, A. | | Deposit date: | 2006-08-13 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Ultrahigh-resolution study of protein atomic displacement parameters at cryotemperatures obtained with a helium cryostat.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
