1HVH
 
 | | NONPEPTIDE CYCLIC CYANOGUANIDINES AS HIV PROTEASE INHIBITORS | | Descriptor: | HIV-1 PROTEASE, {[4-R(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-1,3-BIS(4-HYDROXYMETHYL)METHYL]-4,7-BIS(PHENYLMETHYL) -2H-1,3-DIAZEPIN-2-YLIDENE]CYANAMIDE} | | Authors: | Chang, C.-H. | | Deposit date: | 1997-12-13 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nonpeptide cyclic cyanoguanidines as HIV-1 protease inhibitors: synthesis, structure-activity relationships, and X-ray crystal structure studies.
J.Med.Chem., 41, 1998
|
|
6H7W
 
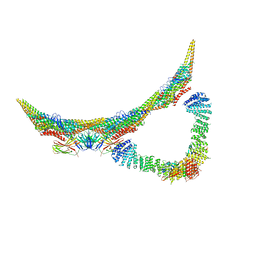 | | Model of retromer-Vps5 complex assembled on membrane. | | Descriptor: | Putative vacuolar protein sorting-associated protein, Vacuolar protein sorting-associated protein 26-like protein, Vacuolar protein sorting-associated protein 29, ... | | Authors: | Kovtun, O, Leneva, N, Ariotti, N, Rohan, T.S, Owen, D.J, Briggs, J.A.G, Collins, B.M. | | Deposit date: | 2018-07-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.4 Å) | | Cite: | Structure of the membrane-assembled retromer coat determined by cryo-electron tomography.
Nature, 561, 2018
|
|
3P8H
 
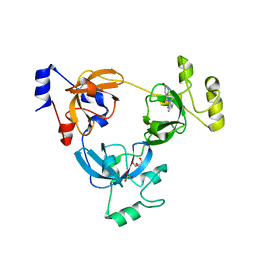 | | Crystal structure of L3MBTL1 (MBT repeat) in complex with a nicotinamide antagonist | | Descriptor: | 3-bromo-5-[(4-pyrrolidin-1-ylpiperidin-1-yl)carbonyl]pyridine, GLYCEROL, Lethal(3)malignant brain tumor-like protein, ... | | Authors: | Lam, R, Herold, J.M, Ouyang, H, Tempel, W, Gao, C, Ravichandran, M, Senisterra, G, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Kireev, D, Frye, S.V, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-13 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small-molecule ligands of methyl-lysine binding proteins.
J.Med.Chem., 54, 2011
|
|
2QZW
 
 | |
2QPW
 
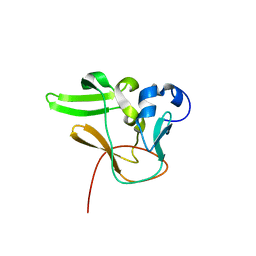 | | Methyltransferase domain of human PR domain-containing protein 2 | | Descriptor: | PR domain zinc finger protein 2 | | Authors: | Lunin, V.V, Wu, H, Dombrovski, L, Antoshenko, T, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-25 | | Release date: | 2007-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
3RQ4
 
 | | Crystal structure of suppressor of variegation 4-20 homolog 2 | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SUV420H2, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Zeng, H, Tempel, W, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-27 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2.
Febs Lett., 587, 2013
|
|
3C0Z
 
 | | Crystal structure of catalytic domain of human histone deacetylase HDAC7 in complex with SAHA | | Descriptor: | Histone deacetylase 7a, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, POTASSIUM ION, ... | | Authors: | Min, J, Schuetz, A, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity.
J.Biol.Chem., 283, 2008
|
|
3MAV
 
 | | Crystal structure of Plasmodium vivax putative farnesyl pyrophosphate synthase (Pv092040) | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION | | Authors: | Dong, A, Dunford, J, Lew, J, Wernimont, A.K, Ren, H, Zhao, Y, Koeieradzki, I, Opperman, U, Sundstrom, M, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular characterization of a novel geranylgeranyl pyrophosphate synthase from Plasmodium parasites.
J.Biol.Chem., 286, 2011
|
|
1PFD
 
 | | THE SOLUTION STRUCTURE OF HIGH PLANT PARSLEY [2FE-2S] FERREDOXIN, NMR, 18 STRUCTURES | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Im, S.-C, Liu, G, Luchinat, C, Sykes, A.G, Bertini, I. | | Deposit date: | 1998-05-05 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of parsley [2Fe-2S]ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
2QZX
 
 | | Secreted aspartic proteinase (Sap) 5 from Candida albicans | | Descriptor: | Candidapepsin-5, Pepstatin | | Authors: | Lee, J.H, Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2007-08-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of Sap1 and Sap5: Structural comparison of the secreted aspartic proteinases from Candida albicans.
Proteins, 72, 2008
|
|
4GKP
 
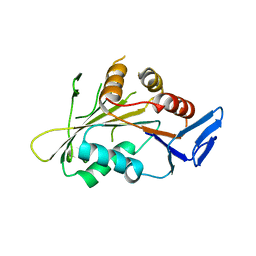 | |
3DEL
 
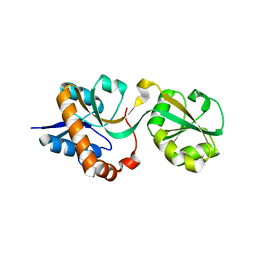 | |
2IGQ
 
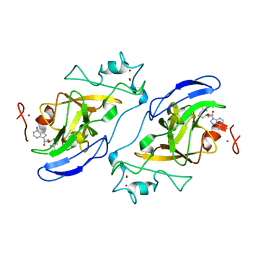 | | Human euchromatic histone methyltransferase 1 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, euchromatic histone methyltransferase 1 | | Authors: | Min, J, Wu, H, Antoshenko, T, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-23 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
1PS2
 
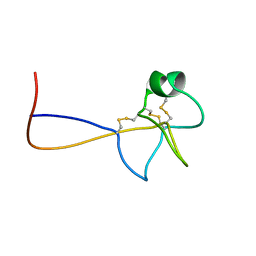 | | HIGH RESOLUTION NMR SOLUTION STRUCTURE OF HUMAN PS2, 19 STRUCTURES | | Descriptor: | PS2 | | Authors: | Williams, M.A, Polshakov, V.I, Gargaro, A.R, Feeney, J. | | Deposit date: | 1997-01-07 | | Release date: | 1997-07-07 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of human pNR-2/pS2: a single trefoil motif protein.
J.Mol.Biol., 267, 1997
|
|
4MI0
 
 | | Human Enhancer of Zeste (Drosophila) Homolog 2(EZH2) | | Descriptor: | Histone-lysine N-methyltransferase EZH2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations.
Plos One, 8, 2013
|
|
1HYM
 
 | | HYDROLYZED TRYPSIN INHIBITOR (CMTI-V, MINIMIZED AVERAGE NMR STRUCTURE) | | Descriptor: | HYDROLYZED CUCURBITA MAXIMA TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Prakash, O, Krishnamoorthi, R. | | Deposit date: | 1995-06-12 | | Release date: | 1995-09-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Reactive-site hydrolyzed Cucurbita maxima trypsin inhibitor-V: function, thermodynamic stability, and NMR solution structure.
Biochemistry, 34, 1995
|
|
3F70
 
 | | Crystal structure of L3MBTL2-H4K20me1 complex | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein, N-METHYL-LYSINE | | Authors: | Guo, Y, Qi, C, Allali-Hassani, A, Pan, P, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Botchkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
3BPR
 
 | | Crystal structure of catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor C52 | | Descriptor: | 2-(2-HYDROXYETHYLAMINO)-6-(3-CHLOROANILINO)-9-ISOPROPYLPURINE, CHLORIDE ION, Proto-oncogene tyrosine-protein kinase MER, ... | | Authors: | Walker, J.R, Huang, X, Finerty Jr, P.J, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the inhibited states of the Mer receptor tyrosine kinase.
J.Struct.Biol., 165, 2009
|
|
3ZG0
 
 | | Crystal structure of ceftaroline acyl-PBP2a from MRSA with non- covalently bound ceftaroline and muramic acid at allosteric site obtained by cocrystallization | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ceftaroline, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZFZ
 
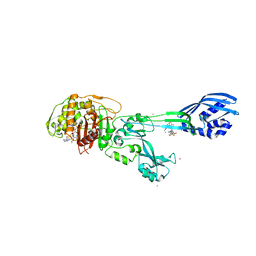 | | Crystal structure of ceftaroline acyl-PBP2a from MRSA with non- covalently bound ceftaroline and muramic acid at allosteric site obtained by soaking | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ceftaroline, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2LT2
 
 | |
3L6J
 
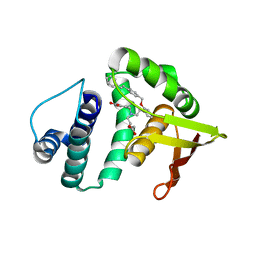 | | Structure of cinaciguat (bay 58-2667) bound to nostoc H-NOX domain | | Descriptor: | 4-({(4-carboxybutyl)[2-(2-{[4-(2-phenylethyl)benzyl]oxy}phenyl)ethyl]amino}methyl)benzoic acid, Alr2278 protein | | Authors: | Martin, F, van den Akker, F. | | Deposit date: | 2009-12-23 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of cinaciguat (BAY 58-2667) bound to Nostoc H-NOX domain reveals insights into heme-mimetic activation of the soluble guanylyl cyclase.
J.Biol.Chem., 285, 2010
|
|
4IHO
 
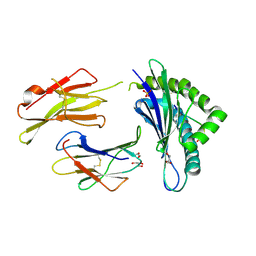 | | Crystal structure of H-2Db Y159F in complex with chimeric gp100 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Uchtenhagen, H, Stahl, E, Achour, A. | | Deposit date: | 2012-12-19 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Proline substitution independently enhances H-2D(b) complex stabilization and TCR recognition of melanoma-associated peptides
Eur.J.Immunol., 43, 2013
|
|
4BL3
 
 | | Crystal structure of PBP2a clinical mutant N146K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, PENICILLIN BINDING PROTEIN 2 PRIME, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Carrasco-Lopez, C, Hermoso, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
3G1Q
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei in ligand free state | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha-demethylase | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Harp, J, Wawrzak, Z, Waterman, M.R, Park, H. | | Deposit date: | 2009-01-30 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Trypanosoma brucei sterol 14alpha-demethylase and implications for selective treatment of human infections.
J.Biol.Chem., 285, 2010
|
|
