6LTC
 
 | |
5M4H
 
 | | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase | | Descriptor: | Heat shock protein HSP 90-alpha, SULFATE ION, [2,4-bis(oxidanyl)phenyl]-[(7~{S})-7-(trifluoromethyl)-6,7-dihydro-5~{H}-pyrazolo[1,5-a]pyrimidin-4-yl]methanone | | Authors: | Baker, L.M, Brough, P, Surgenor, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase.
J. Med. Chem., 60, 2017
|
|
7VF1
 
 | |
5M4M
 
 | | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase | | Descriptor: | CHLORIDE ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, ... | | Authors: | Baker, L.M, Brough, P, Surgenor, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase.
J. Med. Chem., 60, 2017
|
|
7B38
 
 | | Torpedo californica acetylcholinesterase complexed with Mg+2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L, Weiner, L. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
5LZI
 
 | | Porcine heat-labile enterotoxin R13H in complex with inhibitor MM146 | | Descriptor: | (2~{R},4~{S},5~{R},6~{R})-5-acetamido-2-[4-[(2~{R})-3-[2-[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]ethylamino]-3-oxidanylidene-2-(2-phenylethanoylamino)propyl]-1,2,3-triazol-1-yl]-4-oxidanyl-6-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, (2~{R},4~{S},5~{R},6~{R})-5-acetamido-2-[4-[(2~{S})-3-[2-[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]ethylamino]-3-oxidanylidene-2-(2-phenylethanoylamino)propyl]-1,2,3-triazol-1-yl]-4-oxidanyl-6-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Heggelund, J.E, Mackenzie, A, Krengel, U. | | Deposit date: | 2016-09-29 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Towards new cholera prophylactics and treatment: Crystal structures of bacterial enterotoxins in complex with GM1 mimics.
Sci Rep, 7, 2017
|
|
3EFI
 
 | | Carbonic anhydrase activators: Kinetic and X-ray crystallographic study for the interaction of d- and l-tryptophan with the mammalian isoforms I-XIV | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, TRYPTOPHAN, ... | | Authors: | Temperini, C, Innocenti, A, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2008-09-09 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbonic anhydrase activators: kinetic and X-ray crystallographic study for the interaction of D- and L-tryptophan with the mammalian isoforms I-XIV
Bioorg.Med.Chem., 16, 2008
|
|
7B2W
 
 | | Torpedo californica acetylcholinesterase complexed with UO2 | | Descriptor: | Acetylcholinesterase, URANYL (VI) ION | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L, Weiner, L. | | Deposit date: | 2020-11-28 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
7VF8
 
 | | Crystal Structure of HasAp with Co-5-octaethyloxaporphyrinium cation | | Descriptor: | CITRIC ACID, Co-5-octaethyloxaporphyrinium cation, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Takiguchi, A, Sakakibara, E, Sugimoto, H, Shoji, O, Shinokubo, H. | | Deposit date: | 2021-09-10 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of HasAp with Co-5-octaethyloxaporphyrinium cation
To Be Published
|
|
6LTD
 
 | |
7VMF
 
 | |
7ZIE
 
 | |
7VDN
 
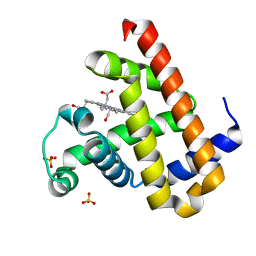 | | High resolution crystal structure of Sperm Whale Myoglobin in the carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shibayama, N, Sato-Tomita, A, Ishimoto, N, Park, S.Y. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | X-ray fluorescence holography of biological metal sites: Application to myoglobin.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
7VF7
 
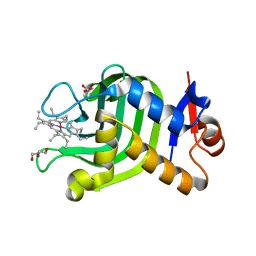 | | Crystal Structure of HasAp with Co-octaethylporphyrin | | Descriptor: | Co-octaethylporphyrin, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Takiguchi, A, Sakakibara, E, Sugimoto, H, Shoji, O, Shinokubo, H. | | Deposit date: | 2021-09-10 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of HasAp with Co-octaethylporphyrin
To Be Published
|
|
1F6N
 
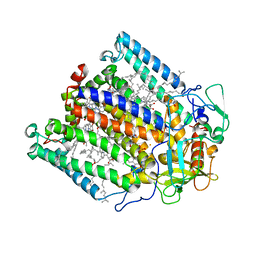 | | CRYSTAL STRUCTURE ANALYSIS OF THE MUTANT REACTION CENTER PRO L209-> TYR FROM THE PHOTOSYNTHETIC PURPLE BACTERIUM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Kuglstatter, A, Ermler, U, Michel, H, Baciou, L, Fritzsch, G. | | Deposit date: | 2000-06-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure analyses of photosynthetic reaction center variants from Rhodobacter sphaeroides: structural changes induced by point mutations at position L209 modulate electron and proton transfer.
Biochemistry, 40, 2001
|
|
4IAU
 
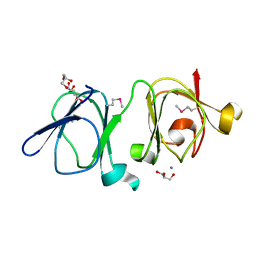 | | Atomic resolution structure of Geodin, a beta-gamma crystallin from Geodia cydonium | | Descriptor: | Beta-gamma-crystallin, CALCIUM ION, GLYCEROL | | Authors: | Vergara, A, Grassi, M, Sica, F, Mazzarella, L, Merlino, A. | | Deposit date: | 2012-12-07 | | Release date: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A novel interdomain interface in crystallins: structural characterization of the [beta][gamma]-crystallin from Geodia cydonium at 0.99 A resolution
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8A29
 
 | | Apo 1-deoxy-D-xylulose 5-phosphate synthase from Pseudomonas aeruginosa | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hamid, R, Adam, S, Lacour, A, Monjas, L, Hirsch, A. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1-deoxy-D-xylulose-5-phosphate synthase from Pseudomonas aeruginosa and Klebsiella pneumoniae reveals conformational changes upon cofactor binding.
J.Biol.Chem., 299, 2023
|
|
1BXY
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L30 FROM THERMUS THERMOPHILUS AT 1.9 A RESOLUTION: CONFORMATIONAL FLEXIBILITY OF THE MOLECULE. | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L30) | | Authors: | Fedorov, R, Nevskaya, N, Khairullina, A, Tishchenko, S, Mikhailov, A, Garber, M, Nikonov, S. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ribosomal protein L30 from Thermus thermophilus at 1.9 A resolution: conformational flexibility of the molecule.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6EX5
 
 | |
5M4P
 
 | | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase | | Descriptor: | CHLORIDE ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, ... | | Authors: | Baker, L.M, Brough, P, Surgenor, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase.
J. Med. Chem., 60, 2017
|
|
2Z5T
 
 | | Molecular basis for the inhibition of p53 by Mdmx | | Descriptor: | Cellular tumor antigen p53, Mdm4 protein | | Authors: | Popowicz, G.M, Czarna, A, Rothweiler, U, Szwagierczak, A, Holak, T.A. | | Deposit date: | 2007-07-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of p53 by Mdmx.
Cell Cycle, 6, 2007
|
|
3II0
 
 | | Crystal structure of human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase, cytoplasmic, ... | | Authors: | Ugochukwu, E, Pilka, E, Cooper, C, Bray, J.E, Yue, W.W, Muniz, J, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human Glutamate oxaloacetate transaminase 1 (GOT1)
To be Published
|
|
2UVK
 
 | | Structure of YjhT | | Descriptor: | YJHT | | Authors: | Muller, A, Severi, E, Wilson, K.S, Thomas, G.H. | | Deposit date: | 2007-03-12 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sialic Acid Mutarotation is Catalyzed by the Escherichia Coli Beta-Propeller Protein Yjht.
J.Biol.Chem., 283, 2008
|
|
6M1C
 
 | |
4HSR
 
 | | Crystal Structure of a class III engineered cephalosporin acylase | | Descriptor: | 5,5-dihydroxy-L-norvaline, glutaryl-7-aminocephalosporanic acid acylase alpha chain, glutaryl-7-aminocephalosporanic acid acylase beta chain | | Authors: | Vrielink, A, Golden, E, Patterson, R, Tie, W.J, Anandan, A, Flematti, G, Molla, G, Rosini, E, Pollegioni, L. | | Deposit date: | 2012-10-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of a class III engineered cephalosporin acylase: comparisons with class I acylase and implications for differences in substrate specificity and catalytic activity.
Biochem.J., 451, 2013
|
|
