3NUK
 
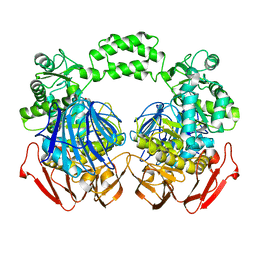 | | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174 | | Descriptor: | ALPHA-GLUCOSIDASE, GLYCEROL | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-07 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE W169Y mutant of ALPHA-GLUCOSIDASE (FAMILY 31) from RUMINOCOCCUS OBEUM ATCC 29174
TO BE PUBLISHED
|
|
1YB2
 
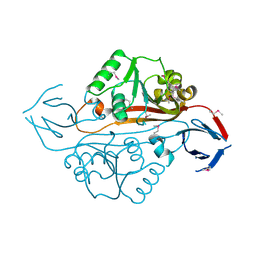 | | Structure of a putative methyltransferase from Thermoplasma acidophilum. | | Descriptor: | hypothetical protein Ta0852 | | Authors: | Cuff, M.E, Xu, X, Edwards, A, Savchenko, A, Sanishvili, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-18 | | Release date: | 2005-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of a putative methyltransferase from Thermoplasma acidophilum.
To be Published
|
|
1Y9B
 
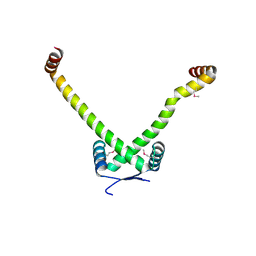 | | Structure of Conserved Putative Transcriptional Factor from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | conserved hypothetical protein | | Authors: | Binkowski, T.A, Hatzos, C, Quartey, P, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-15 | | Release date: | 2005-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conserverd hypothetical protein from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3D1L
 
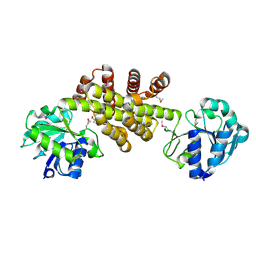 | | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis | | Descriptor: | 2-MERCAPTO-PROPION ALDEHYDE, CHLORIDE ION, Putative NADP oxidoreductase BF3122 | | Authors: | Chang, C, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of putative NADP oxidoreductase BF3122 from Bacteroides fragilis.
To be Published
|
|
4KUN
 
 | | Crystal structure of Legionella pneumophila Lpp1115 / KaiB | | Descriptor: | Hypothetical protein Lpp1115 | | Authors: | Petit, P, Stogios, P.J, Stein, A, Wawrzak, Z, Skarina, T, Daniels, C, Di Leo, R, Buchrieser, C, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-22 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Legionella pneumophila kai operon is implicated in stress response and confers fitness in competitive environments.
Environ Microbiol, 16, 2014
|
|
3CJN
 
 | | Crystal structure of transcriptional regulator, MarR family, from Silicibacter pomeroyi | | Descriptor: | PHOSPHATE ION, Transcriptional regulator, MarR family | | Authors: | Chang, C, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-13 | | Release date: | 2008-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of MarR family transcriptional regulator from Silicibacter pomeroyi.
To be Published
|
|
3CTV
 
 | | Crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, SULFATE ION | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-14 | | Release date: | 2008-04-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | X-ray crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus.
To be Published
|
|
1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3D1P
 
 | | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative thiosulfate sulfurtransferase YOR285W | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae.
To be Published
|
|
2LF6
 
 | | Solution NMR structure of HopABPph1448_220_320 from Pseudomonas syringae pv. phaseolicola str. 1448A, Midwest Center for Structural Genomics target APC40132.4 and Northeast Structural Genomics Consortium target PsT3A | | Descriptor: | Effector protein hopAB1 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
3O60
 
 | |
3O12
 
 | | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein YJL217W | | Authors: | Zhang, R, Tan, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae.
TO BE PUBLISHED
|
|
3OCJ
 
 | | The crystal structure of a possilbe exported protein from Bordetella parapertussis | | Descriptor: | GLYCEROL, PALMITIC ACID, Putative exported protein | | Authors: | Tan, K, Bigelow, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The crystal structure of a possilbe exported protein from Bordetella parapertussis
To be Published
|
|
1MKZ
 
 | | Crystal structure of MoaB protein at 1.6 A resolution. | | Descriptor: | ACETIC ACID, Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Sanishvili, R, Skarina, T, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Escherichia coli MoaB suggests a probable role in molybdenum cofactor synthesis.
J.Biol.Chem., 279, 2004
|
|
3HIM
 
 | | The Crystal Structure of a Bacterial Regulatory Protein in the tetR Family from Rhodococcus RHA1 to 2.2A | | Descriptor: | Probable transcriptional regulator | | Authors: | Stein, A.J, Binkowski, T.A, Evdokimova, E, Kagan, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of a Bacterial Regulatory Protein in the tetR Family from Rhodococcus RHA1 to 2.2A
To be Published
|
|
1MKI
 
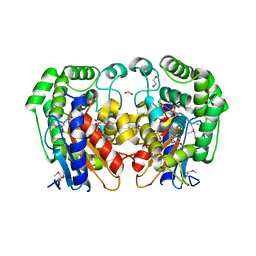 | | Crystal Structure of Bacillus Subtilis Probable Glutaminase, APC1040 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable Glutaminase ybgJ | | Authors: | Kim, Y, Dementieva, I, Vinokour, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
3OHR
 
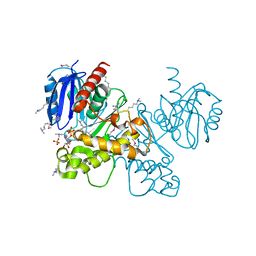 | | Crystal structure of fructokinase from bacillus subtilis complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative fructokinase, SULFATE ION, ... | | Authors: | Nocek, B, Volkart, L, Cuff, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural studies of ROK fructokinase YdhR from Bacillus subtilis: insights into substrate binding and fructose specificity.
J.Mol.Biol., 406, 2011
|
|
3OLQ
 
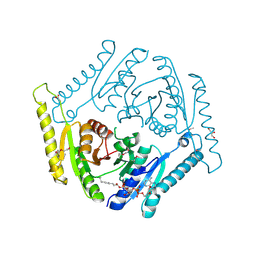 | | The crystal structure of a universal stress protein E from Proteus mirabilis HI4320 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | The crystal structure of a universal stress protein E from
Proteus mirabilis HI4320
To be Published
|
|
3ON2
 
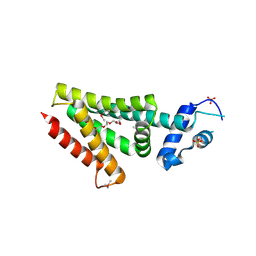 | | Structure of a protein with unknown function from Rhodococcus sp. RHA1 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Probable transcriptional regulator, SULFATE ION | | Authors: | Fan, Y, Evdokimova, E, Egorova, O, Savchenko, A, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of a protein with unknown function from Rhodococcus sp. RHA1
To be Published
|
|
1I36
 
 | | Structure of Conserved Protein MTH1747 of Unknown Function Reveals Structural Similarity with 3-Hydroxyacid Dehydrogenases | | Descriptor: | CONSERVED HYPOTHETICAL PROTEIN MTH1747, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korolev, S.V, Dementieva, I.S, Christendat, D, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-02-13 | | Release date: | 2002-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURAL SIMILARITIES OF MTH1747 HYPOTHETICAL PROTEIN FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH 3-HYDROXYACID DEHYDROGENASES
to be published
|
|
1NPY
 
 | | Structure of shikimate 5-dehydrogenase-like protein HI0607 | | Descriptor: | ACETYL GROUP, Hypothetical shikimate 5-dehydrogenase-like protein HI0607 | | Authors: | Korolev, S, Koroleva, O, Zarembinski, T, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of a Novel Shikimate Dehydrogenase from Haemophilus influenzae.
J.Biol.Chem., 280, 2005
|
|
1NJH
 
 | |
1NC7
 
 | | Crystal Structure of Thermotoga maritima 1070 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima Hypothetical protein TM1070
To be Published
|
|
3PGP
 
 | | Crystal structure of PA4794 - GNAT superfamily protein in complex with AcCoA | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
3SOZ
 
 | | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2 | | Descriptor: | Cytoplasmic Protein STM1381, GLYCEROL | | Authors: | Joachimiak, A, Duke, N.E.C, Jedrzejczak, R, Li, H, Adkins, J, Brown, R, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cytoplasmic Protein STM1381 from Salmonella typhimurium LT2
To be Published
|
|
