5CMZ
 
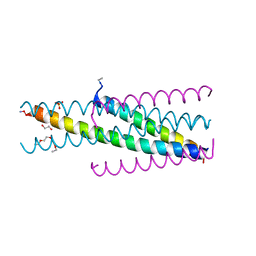 | | Artificial HIV fusion inhibitor AP3 fused to the C-terminus of gp41 NHR | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Artificial HIV entry inhibitor AP3, ... | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CN0
 
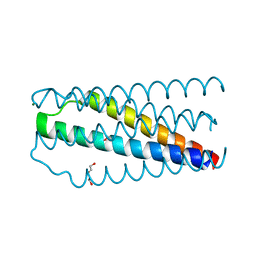 | | Artificial HIV fusion inhibitor AP2 fused to the C-terminus of gp41 NHR | | Descriptor: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein,AP2, MAGNESIUM ION | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CMU
 
 | |
7YIK
 
 | |
7YIL
 
 | |
7YKV
 
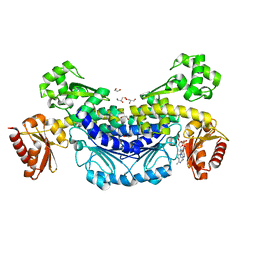 | | The structure of archaeal nuclease RecJ2 from Methanocaldococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, [5-(6-aminopurin-9-yl)-2-(hydroxymethyl)furan-3-yl] [5-(6-aminopurin-9-yl)-3-oxidanyl-furan-2-yl]methyl hydrogen phosphate, ... | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-07-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The structure of archaeal nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
7YOQ
 
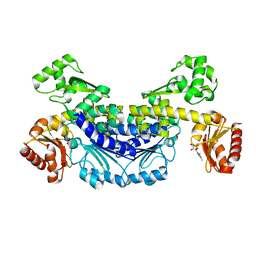 | | Recj2 and CMP complex from Methanocaldococcus jannaschii | | Descriptor: | [5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)furan-2-yl]methyl dihydrogen phosphate, nuclease RecJ2 | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | The structure of nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
7YOS
 
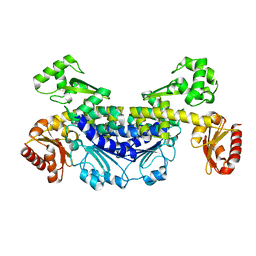 | |
7YOR
 
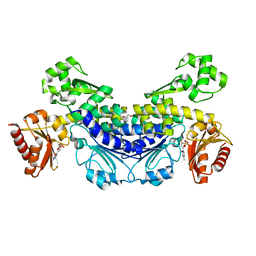 | | Recj2 and dCMP complex from Methanocaldococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, [5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-furan-2-yl]methyl dihydrogen phosphate, archaeal nuclease RecJ2 | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structure of nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
6KXS
 
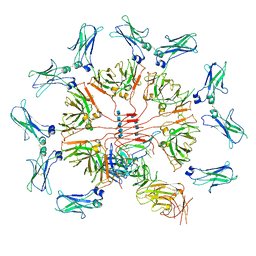 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the ectodomain of pIgR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Li, Y, Wang, G, Xiao, J. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-05 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into immunoglobulin M.
Science, 367, 2020
|
|
6NKH
 
 | |
3T0H
 
 | |
3T0Z
 
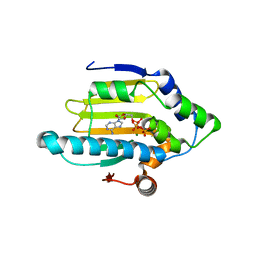 | | Hsp90 N-terminal domain bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
6NKK
 
 | | Structure of PhqE Reductase/Diels-Alderase from Penicillium fellutanum in complex with NADP+ and premalbrancheamide | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Newmister, S.A, Dan, Q, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
6LZ7
 
 | |
6NKI
 
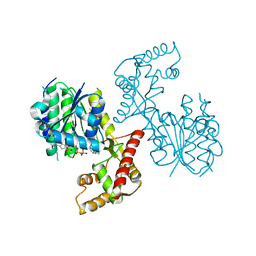 | | Structure of PhqB Reductase Domain from Penicillium fellutanum | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NRPS | | Authors: | Dan, Q, Newmister, S.A, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
6LTK
 
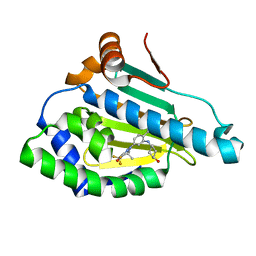 | | HSP90 in complex with SNX-2112 | | Descriptor: | 4-[6,6-dimethyl-4-oxidanylidene-3-(trifluoromethyl)-5,7-dihydroindazol-1-yl]-2-[(4-oxidanylcyclohexyl)amino]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Complex Crystal Structure Determination and in vitro Anti-non-small Cell Lung Cancer Activity of Hsp90 N Inhibitor SNX-2112.
Front Cell Dev Biol, 9, 2021
|
|
6LZ3
 
 | |
6NKM
 
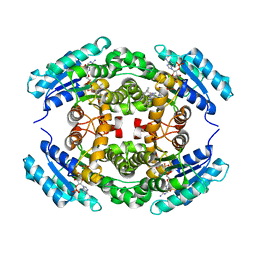 | | Structure of PhqE D166N Reductase/Diels-Alderase from Penicillium fellutanum in complex with NADP+ and substrate | | Descriptor: | 3-{[2-(2-methylbut-3-en-2-yl)-1H-indol-3-yl]methyl}-8H-pyrrolo[1,2-a]pyrazin-5-ium-1-olate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Newmister, S.A, Dan, Q, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
3T10
 
 | | HSP90 N-terminal domain bound to ACP | | Descriptor: | Heat shock protein HSP 90-alpha, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
4M57
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 from maize | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10 | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
4M59
 
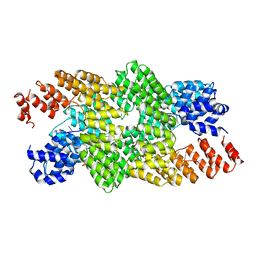 | | Crystal structure of the pentatricopeptide repeat protein PPR10 in complex with an 18-nt psaJ RNA element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
5GUF
 
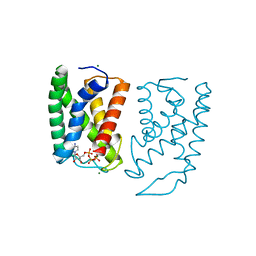 | |
4NJL
 
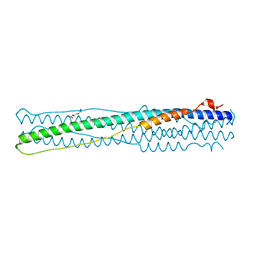 | | Crystal structure of middle east respiratory syndrome coronavirus S2 protein fusion core | | Descriptor: | S protein, TRIETHYLENE GLYCOL | | Authors: | Zhu, Y, Lu, L, Qin, L, Ye, S, Jiang, S, Zhang, R. | | Deposit date: | 2013-11-10 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor.
Nat Commun, 5, 2014
|
|
5H0N
 
 | |
