7LJR
 
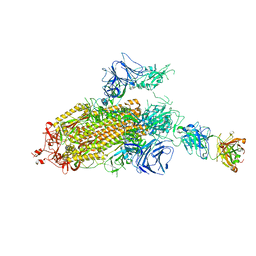 | | SARS-CoV-2 Spike Protein Trimer bound to DH1043 fab | | Descriptor: | Fab DH1043 heavy chain, Fab DH1043 light chain, Spike glycoprotein | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates.
Biorxiv, 2021
|
|
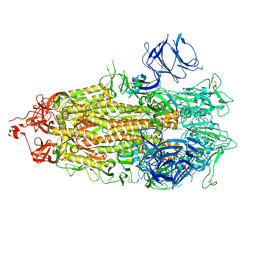
7LWS
 
 | |
7LG6
 
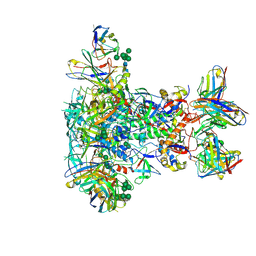 | | BG505 SOSIP.v5.2 in complex with VRC40.01 and RM19R Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, Torres, J.L, Wu, N.R, Ward, A.B. | | Deposit date: | 2021-01-19 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis of glycan276-dependent recognition by HIV-1 broadly neutralizing antibodies.
Cell Rep, 37, 2021
|
|
3JDW
 
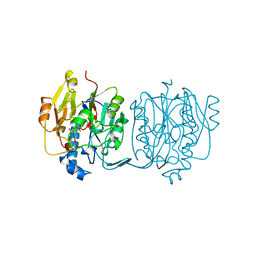 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | L-ARGININE:GLYCINE AMIDINOTRANSFERASE, L-ornithine | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
1QU0
 
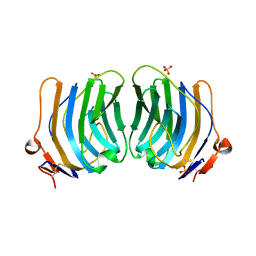 | | CRYSTAL STRUCTURE OF THE FIFTH LAMININ G-LIKE MODULE OF THE MOUSE LAMININ ALPHA2 CHAIN | | Descriptor: | CALCIUM ION, LAMININ ALPHA2 CHAIN, SULFATE ION | | Authors: | Hohenester, E, Tisi, D, Talts, J.F, Timpl, R. | | Deposit date: | 1999-07-05 | | Release date: | 1999-12-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of a laminin G-like module reveals the molecular basis of alpha-dystroglycan binding to laminins, perlecan, and agrin.
Mol.Cell, 4, 1999
|
|
1I77
 
 | | CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ESSEX 6 | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Einsle, O, Foerster, S, Mann, K.H, Fritz, G, Messerschmidt, A, Kroneck, P.M.H. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Spectroscopic investigation and determination of reactivity and structure of the tetraheme cytochrome c3 from Desulfovibrio desulfuricans Essex 6.
Eur.J.Biochem., 268, 2001
|
|
1AW9
 
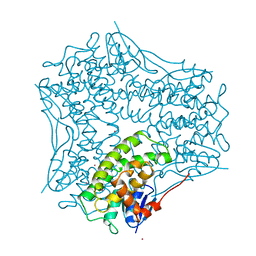 | | STRUCTURE OF GLUTATHIONE S-TRANSFERASE III IN APO FORM | | Descriptor: | CADMIUM ION, GLUTATHIONE S-TRANSFERASE III | | Authors: | Neuefeind, T, Huber, R, Reinemer, P, Knaeblein, J. | | Deposit date: | 1997-10-13 | | Release date: | 1998-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, sequencing, crystallization and X-ray structure of glutathione S-transferase-III from Zea mays var. mutin: a leading enzyme in detoxification of maize herbicides.
J.Mol.Biol., 274, 1997
|
|
1BQB
 
 | | AUREOLYSIN, STAPHYLOCOCCUS AUREUS METALLOPROTEINASE | | Descriptor: | CALCIUM ION, PROTEIN (AUREOLYSIN), ZINC ION | | Authors: | Medrano, F.J, Banbula, A, Potempa, J, Travis, J, Bode, W. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Amino-acid sequence and three-dimensional structure of the Staphylococcus aureus metalloproteinase at 1.72 A resolution.
Structure, 6, 1998
|
|
1DY0
 
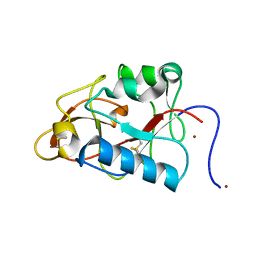 | |
1DY1
 
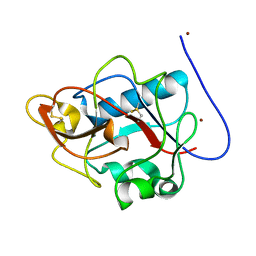 | |
1FD9
 
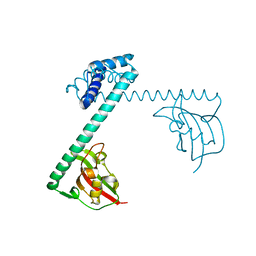 | | CRYSTAL STRUCTURE OF THE MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN (MIP) A MAJOR VIRULENCE FACTOR FROM LEGIONELLA PNEUMOPHILA | | Descriptor: | PROTEIN (MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN), ZINC ION | | Authors: | Riboldi-Tunnicliffe, A, Jessen, S, Konig, B, Rahfeld, J, Hacker, J, Fischer, G, Hilgenfeld, R. | | Deposit date: | 2000-07-20 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of Mip, a prolylisomerase from Legionella pneumophila
Nat.Struct.Biol., 8, 2001
|
|
1KLO
 
 | |
1TLE
 
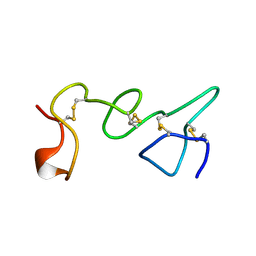 | | LE (LAMININ-TYPE EGF-LIKE) MODULE GIII4 IN SOLUTION AT PH 3.5 AND 290 K, NMR, 14 STRUCTURES | | Descriptor: | LAMININ | | Authors: | Baumgartner, R, Czisch, M, Mayer, U, Schl, E.P, Huber, R, Timpl, R, Holak, T.A. | | Deposit date: | 1996-01-26 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the nidogen binding LE module of the laminin gamma1 chain in solution.
J.Mol.Biol., 257, 1996
|
|
3ZGG
 
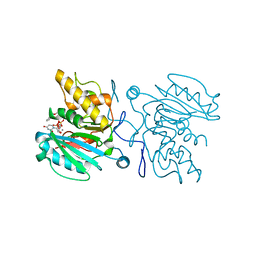 | | Crystal structure of the Fucosylgalactoside alpha N- acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with NPE caged UDP-Gal (C222(1) space group) | | Descriptor: | 1-(2-NITROPHENYL)ETHYL UDP-GALACTOSE, GLYCEROL, HISTO-BLOOD GROUP ABO SYSTEM TRANSFERASE, ... | | Authors: | Jorgensen, R, Batot, G.O, Hindsgaul, O, Tanaka, H, Perez, S, Imberty, A, Breton, C, Royant, A, Palcic, M.M. | | Deposit date: | 2012-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of a Human Blood Group Glycosyltransferase in Complex with a Photo-Activatable Udp-Gal Derivative Reveal Two Different Binding Conformations
Acta Crystallogr.,Sect.F, 70, 2014
|
|
2JDW
 
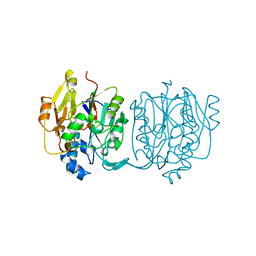 | |
1OKQ
 
 | | LAMININ ALPHA 2 CHAIN LG4-5 DOMAIN PAIR, CA1 SITE MUTANT | | Descriptor: | CALCIUM ION, LAMININ ALPHA 2 CHAIN | | Authors: | Wizemann, H, Garbe, J.H.O, Friedrich, M.V.K, Timpl, R, Sasaki, T, Hohenester, E. | | Deposit date: | 2003-07-28 | | Release date: | 2003-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distinct Requirements for Heparin and Alpha-Dystroglycan Binding Revealed by Structure-Based Mutagenesis of the Laminin Alpha2 Lg4-Lg5 Domain Pair
J.Mol.Biol., 332, 2003
|
|
1JDW
 
 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | BETA-MERCAPTOETHANOL, L-ARGININE:GLYCINE AMIDINOTRANSFERASE | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-22 | | Release date: | 1998-01-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
1SPP
 
 | | THE CRYSTAL STRUCTURES OF TWO MEMBERS OF THE SPERMADHESIN FAMILY REVEAL THE FOLDING OF THE CUB DOMAIN | | Descriptor: | MAJOR SEMINAL PLASMA GLYCOPROTEIN PSP-I, MAJOR SEMINAL PLASMA GLYCOPROTEIN PSP-II | | Authors: | Romero, A, Romao, M.J, Varela, P.F, Kolln, I, Dias, J.M, Carvalho, A.L, Sanz, L, Topfer-Petersen, E, Calvete, J.J. | | Deposit date: | 1997-06-19 | | Release date: | 1998-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of two spermadhesins reveal the CUB domain fold.
Nat.Struct.Biol., 4, 1997
|
|
4JDW
 
 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | ARGININE, L-ARGININE:GLYCINE AMIDINOTRANSFERASE | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
1DYK
 
 | | Laminin alpha 2 chain LG4-5 domain pair | | Descriptor: | CALCIUM ION, LAMININ ALPHA 2 CHAIN | | Authors: | Tisi, D, Talts, J.F, Timple, R, Hohenester, E. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-Terminal Laminin G-Like Domain Pair of the Laminin Alpha 2 Chain Harbouring Binding Sites for Alpha-Dystroglycan and Heparin
Embo J., 19, 2000
|
|
1GL4
 
 | | Nidogen-1 G2/Perlecan IG3 Complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BASEMENT MEMBRANE-SPECIFIC HEPARAN SULFATE PROTEOGLYCAN CORE PROTEIN, NIDOGEN-1, ... | | Authors: | Kvansakul, M, Hopf, M, Ries, A, Timpl, R, Hohenester, E. | | Deposit date: | 2001-08-23 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the High-Affinity Interaction of Nidogen-1 with Immunoglobulin-Like Domain 3 of Perlecan
Embo J., 20, 2001
|
|
1GZ2
 
 | |
1H4U
 
 | | Domain G2 of mouse nidogen-1 | | Descriptor: | NIDOGEN-1 | | Authors: | Hopf, M, Gohring, W, Ries, A, Timpl, R, Hohenester, E. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure and Mutational Analysis of a Perlecan-Binding Fragment of Nidogen-1
Nat.Struct.Biol., 8, 2001
|
|
3ZGF
 
 | | Crystal structure of the Fucosylgalactoside alpha N- acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with in complex with NPE caged UDP-Gal (P2(1)2(1)2(1) space group) | | Descriptor: | 1-(2-NITROPHENYL)ETHYL UDP-GALACTOSE, HISTO-BLOOD GROUP ABO SYSTEM TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Jorgensen, R, Batot, G.O, Hindsgaul, O, Tanaka, H, Perez, S, Imberty, A, Breton, C, Royant, A, Palcic, M.M. | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structures of a Human Blood Group Glycosyltransferase in Complex with a Photo-Activatable Udp-Gal Derivative Reveal Two Different Binding Conformations
Acta Crystallogr.,Sect.F, 70, 2014
|
|
