8W1Y
 
 | | 2.30 angstrom resolution intermediate crystal structure of KatG from Mycobacterium tuberculosis with an MYW-OOH cofactor soaked with peroxide for 1 minute | | Descriptor: | Catalase-peroxidase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, J, Duan, R, Liu, A. | | Deposit date: | 2024-02-19 | | Release date: | 2024-10-23 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Indole N-Linked Hydroperoxyl Adduct of Protein-Derived Cofactor Modulating Catalase-Peroxidase Functions.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
3FZB
 
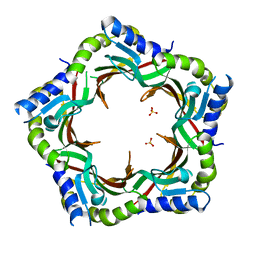 | | Crystal structure of the tail terminator protein from phage lambda (gpU-WT) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-24 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
2L0K
 
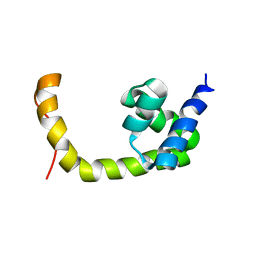 | | NMR solution structure of a transcription factor SpoIIID in complex with DNA | | Descriptor: | Stage III sporulation protein D | | Authors: | Chen, B, Himes, P, Lu, Z, Liu, A, Yan, H, Kroos, L. | | Deposit date: | 2010-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Novel Mode of DNA Binding by Bacterial Transcription Factor SpoIIID
To be Published
|
|
1QM2
 
 | |
1QM1
 
 | |
1QM3
 
 | |
1QLX
 
 | | Human prion protein | | Descriptor: | PRION PROTEIN | | Authors: | Zahn, R, Liu, A, Luhrs, T, Wuthrich, K. | | Deposit date: | 1999-09-17 | | Release date: | 1999-12-16 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1QM0
 
 | |
1QLZ
 
 | | Human prion protein | | Descriptor: | PRION PROTEIN | | Authors: | Zahn, R, Liu, A, Luhrs, T, Wuthrich, K. | | Deposit date: | 1999-09-20 | | Release date: | 1999-12-16 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
8CZP
 
 | |
6MGT
 
 | | Crystal structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase Mutant H110A | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Yang, Y, Daivs, I, Matsui, T, Rubalcava, I, Liu, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Quaternary structure of alpha-amino-beta-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) controls its activity.
J.Biol.Chem., 294, 2019
|
|
6MGS
 
 | | Crystal structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase with Space Group of C2221 | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Yang, Y, Davis, I, Matsui, T, Rubalcava, I, Liu, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.131 Å) | | Cite: | Quaternary structure of alpha-amino-beta-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) controls its activity.
J.Biol.Chem., 294, 2019
|
|
6ON3
 
 | | A substrate bound structure of L-DOPA dioxygenase from Streptomyces sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4-DIHYDROXYPHENYLALANINE, FE (II) ION, ... | | Authors: | Wang, Y, Shin, I, Fu, Y, Colabroy, K, Liu, A. | | Deposit date: | 2019-04-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structures of L-DOPA Dioxygenase fromStreptomyces sclerotialus.
Biochemistry, 58, 2019
|
|
7KQ2
 
 | |
7KPZ
 
 | |
5WP2
 
 | | 1.44 Angstrom crystal structure of CYP121 from Mycobacterium tuberculosis in complex with substrate and CN | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, CYANIDE ION, Mycocyclosin synthase, ... | | Authors: | Fielding, A, Dornevil, K, Liu, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Probing Ligand Exchange in the P450 Enzyme CYP121 from Mycobacterium tuberculosis: Dynamic Equilibrium of the Distal Heme Ligand as a Function of pH and Temperature.
J. Am. Chem. Soc., 139, 2017
|
|
8VYY
 
 | |
7KQR
 
 | | A 1.89-A resolution substrate-bound crystal structure of heme-dependent tyrosine hydroxylase from S. sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heme-dependent L-tyrosine hydroxylase, ... | | Authors: | Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQS
 
 | | A 1.68-A resolution 3-fluoro-L-tyrosine bound crystal structure of heme-dependent tyrosine hydroxylase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQU
 
 | | A 1.58-A resolution crystal structure of ferric-hydroperoxo intermediate of L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Davis, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQT
 
 | |
2HBX
 
 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
2HBV
 
 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
4ERG
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, FE (III) ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4EPK
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6009 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
