9L3Z
 
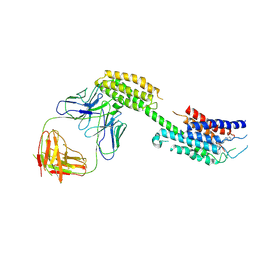 | | Cryo-EM structure of the inactive chemokine-like receptor 1 (CMKLR1) | | Descriptor: | Chemerin-like receptor 1,Soluble cytochrome b562, LRH7-C2, The heavy chain of anti-BRIL Fab, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
9L3W
 
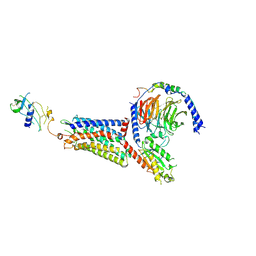 | | Cryo-EM structure of the chemokine-like receptor 1 in complex with chemerin and Gi1 | | Descriptor: | Chemerin-like receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
9L3Y
 
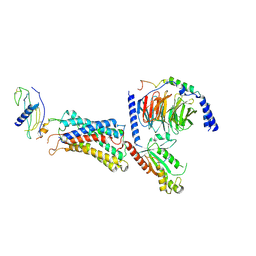 | | Cryo-EM structure of the G-protein coupled receptor 1 (GPR1) in complex with chemerin and Gi1 | | Descriptor: | Chemerin-like receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
6LI3
 
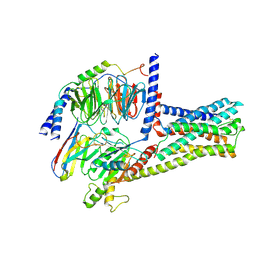 | | cryo-EM structure of GPR52-miniGs-NB35 | | Descriptor: | G-protein coupled receptor 52, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, M, Wang, N, Xu, F, Wu, J, Lei, M. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
9MME
 
 | | ROOLfirm-octamer-wild type | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (523-MER) | | Authors: | Ling, X.B, Ma, J.B, Fang, W.W, Korostelev, A.A. | | Deposit date: | 2024-12-20 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Cryo-EM structure of a natural RNA nanocage.
Nature, 2025
|
|
9MM6
 
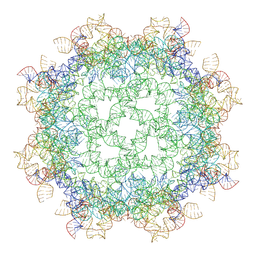 | | ROOLefa-octamer-wild type | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (526-MER) | | Authors: | Ling, X.B, Fang, W.W, Ma, J.B, Korostelev, A.A. | | Deposit date: | 2024-12-20 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structure of a natural RNA nanocage.
Nature, 2025
|
|
9MMG
 
 | |
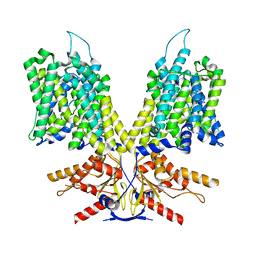
8UC1
 
 | |
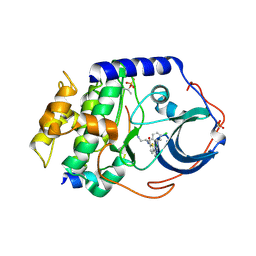
2GU8
 
 | |
7LAT
 
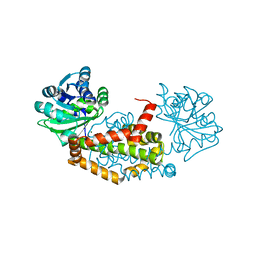 | |
7KH7
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex with Mg2+, NADPH, and NSC116565 | | Descriptor: | 6-hydroxy-2-methyl[1,3]thiazolo[4,5-d]pyrimidine-5,7(4H,6H)-dione, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Kurz, J.L, Patel, K.P, Guddat, L.W. | | Deposit date: | 2020-10-20 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Discovery of a Pyrimidinedione Derivative with Potent Inhibitory Activity against Mycobacterium tuberculosis Ketol-Acid Reductoisomerase.
Chemistry, 27, 2021
|
|
7KE2
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex with Mg2+ and NSC116565 | | Descriptor: | 6-hydroxy-2-methyl[1,3]thiazolo[4,5-d]pyrimidine-5,7(4H,6H)-dione, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION | | Authors: | Kurz, J.L, Patel, K.P, Guddat, L.W. | | Deposit date: | 2020-10-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Discovery of a Pyrimidinedione Derivative with Potent Inhibitory Activity against Mycobacterium tuberculosis Ketol-Acid Reductoisomerase.
Chemistry, 27, 2021
|
|
8X9M
 
 | | Cte-Branch Product, Mg, post | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (808-MER) | | Authors: | Ling, X.B, Ma, J.B. | | Deposit date: | 2023-11-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structures of a natural circularly permuted group II intron reveal mechanisms of branching and backsplicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8X9N
 
 | | Cte_A695U_Branch Intermediate,Mg,inactive | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (219-mer), ... | | Authors: | Ling, X.B, Ma, J.B. | | Deposit date: | 2023-11-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of a natural circularly permuted group II intron reveal mechanisms of branching and backsplicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8X9Q
 
 | | Cte-A695U-branching,Mg,active | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (809-MER) | | Authors: | Ling, X.B, Ma, J.B. | | Deposit date: | 2023-11-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of a natural circularly permuted group II intron reveal mechanisms of branching and backsplicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8X9O
 
 | | Cte-U537ins-Branching,Ca,inactive | | Descriptor: | CALCIUM ION, POTASSIUM ION, RNA (811-mer) | | Authors: | Ling, X.B, Ma, J.B. | | Deposit date: | 2023-11-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structures of a natural circularly permuted group II intron reveal mechanisms of branching and backsplicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8X9L
 
 | | Cte-U537ins_Branch Intermediate,Ca,active | | Descriptor: | CALCIUM ION, POTASSIUM ION, RNA (219-MER), ... | | Authors: | Ling, X.B, Ma, J.B. | | Deposit date: | 2023-11-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of a natural circularly permuted group II intron reveal mechanisms of branching and backsplicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8X9K
 
 | | Cte_Branching,Ca,inactive | | Descriptor: | CALCIUM ION, POTASSIUM ION, RNA (808-MER) | | Authors: | Ling, X.B, Ma, J.B. | | Deposit date: | 2023-11-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structures of a natural circularly permuted group II intron reveal mechanisms of branching and backsplicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
4V10
 
 | | Skelemin Association with alpha2b,beta3 Integrin: A Structural Model | | Descriptor: | MYOMESIN-1 | | Authors: | Gorbatyuk, V, Deshmukh, L, Nguyen, K, Vinogradova, O. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Skelemin Association with Alphaiibbeta3 Integrin: A Structural Model.
Biochemistry, 53, 2014
|
|
5X8G
 
 | |
4WAF
 
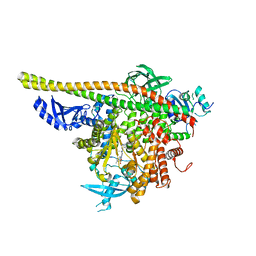 | | Crystal Structure of a novel tetrahydropyrazolo[1,5-a]pyrazine in an engineered PI3K alpha | | Descriptor: | N,N-dimethyl-4-[(6R)-6-methyl-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-3-yl]benzenesulfonamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-Based Drug Design of Novel Potent and Selective Tetrahydropyrazolo[1,5-a]pyrazines as ATR Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4U93
 
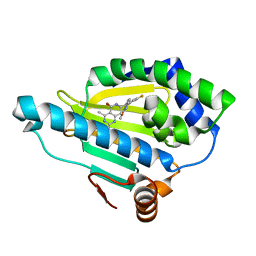 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7R)-2-amino-7-[4-fluoro-2-(6-methoxypyridin-2-yl)phenyl]-4-methyl-7,8-dihydropyrido[4,3-d]pyrimidin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
4W7T
 
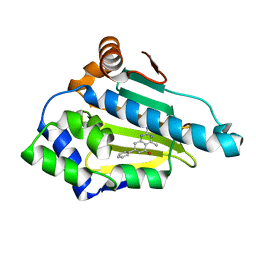 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7S)-2-amino-4-methyl-7-phenyl-7,8-dihydroquinazolin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-22 | | Release date: | 2014-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
6EZP
 
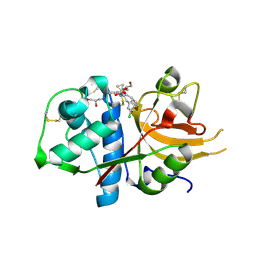 | | CATHEPSIN L IN COMPLEX WITH (3S,14E)-19-chloro-N-(1-cyanocyclopropyl)-5-oxo-12,17-dioxa-4-azatricyclo[16.2.2.06,11]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide | | Descriptor: | (3~{S},14~{E})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, Cathepsin L1, GLYCEROL | | Authors: | Banner, D.W, Benz, J, Kuglstatter, A. | | Deposit date: | 2017-11-16 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EXO
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S},14~{E})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
