8CE8
 
 | | Cytochrome c maturation complex CcmABCDE | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Cytochrome c-type biogenesis protein CcmE, Heme exporter protein B, ... | | Authors: | Ilcu, L, Zhang, L, Einsle, O. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation.
Nat Commun, 14, 2023
|
|
4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
3CJN
 
 | | Crystal structure of transcriptional regulator, MarR family, from Silicibacter pomeroyi | | Descriptor: | PHOSPHATE ION, Transcriptional regulator, MarR family | | Authors: | Chang, C, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-13 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of MarR family transcriptional regulator from Silicibacter pomeroyi.
To be Published
|
|
3CWI
 
 | | Crystal structure of thiamine biosynthesis protein (ThiS) from Geobacter metallireducens. Northeast Structural Genomics Consortium Target GmR137 | | Descriptor: | Thiamine-biosynthesis protein ThiS | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, L, Janjua, H, Xiao, R, Maglaqui, M, Ciccosanti, C, Foote, E.L, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-04-21 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of thiamine biosynthesis protein (ThiS) from Geobacter metallireducens.
To be Published
|
|
5DSF
 
 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-09-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
3CON
 
 | | Crystal structure of the human NRAS GTPase bound with GDP | | Descriptor: | GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Shen, L, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal structure of the human NRAS GTPase bound with GDP.
To be Published
|
|
4UUQ
 
 | | Crystal structure of human mono-glyceride lipase in complex with SAR127303 | | Descriptor: | 4-({[(4-chlorophenyl)sulfonyl]amino}methyl)piperidine-1-carboxylic acid, MONOGLYCERIDE LIPASE | | Authors: | Griebel, G, Pichat, P, Beeske, S, Leroy, T, Redon, N, Francon, D, Bert, L, Even, L, Lopez-Grancha, M, Tolstykh, T, Sun, F, Yu, Q, Brittain, S, Arlt, H, He, T, Zhang, B, Wiederschain, D, Bertrand, T, Houtman, J, Rak, A, Vallee, F, Michot, N, Auge, F, Menet, V, Bergis, O.E, George, P, Avenet, P, Mikol, V, Didier, M, Escoubet, J. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Selective Blockade of the Hydrolysis of the Endocannabinoid 2-Arachidonoylglycerol Impairs Learning and Memory Performance While Producing Antinociceptive Activity in Rodents.
Sci.Rep., 5, 2015
|
|
7SI2
 
 | |
1S13
 
 | | Human Heme Oxygenase Oxidatition of alpha- and gamma-meso-Phenylhemes | | Descriptor: | 2-PHENYLHEME, Heme oxygenase 1 | | Authors: | Wang, J, Niemevz, F, Lad, L, Buldain, G, Poulos, T.L, Ortiz de Montellano, P.R. | | Deposit date: | 2004-01-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Human heme oxygenase oxidation of 5- and 15-phenylhemes.
J.Biol.Chem., 279, 2004
|
|
3CEW
 
 | | Crystal structure of a cupin protein (BF4112) from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR205 | | Descriptor: | Uncharacterized cupin protein, ZINC ION | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Mao, L, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Wang, H, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of a cupin protein (BF4112) from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR205.
To be Published
|
|
3CNW
 
 | | Three-dimensional structure of the protein XoxI (Q81AY6) from Bacillus cereus. Northeast Structural Genomics Consortium target BcR196. | | Descriptor: | Protein XoxI | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, H, Ciccosanti, C, Mao, L, Xiao, R, Nair, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Three-dimensional structure of the protein XoxI (Q81AY6) from Bacillus cereus. Northeast Structural Genomics Consortium target BcR196.
To Be Published
|
|
7OCJ
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS2(Nb14509) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, NbSOS2 (14509) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2021-04-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
4V0G
 
 | | JAK3 in complex with a covalent EGFR inhibitor | | Descriptor: | N-[3-(2-{3-amino-6-[1-(1-methylpiperidin-4-yl)-1H-pyrazol-4-yl]pyrazin-2-yl}-1H-benzimidazol-1-yl)phenyl]propanamide, TYROSINE-PROTEIN KINASE JAK3 | | Authors: | Debreczeni, J.E, Hennessy, E.J, Chuaquini, C, Ashton, S, Coclough, N, Cross, D.A.E, Eberlein, C, Gingipalli, L, Klinowska, T.C.M, Orme, J.P, Sha, L, Wu, X. | | Deposit date: | 2014-09-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Utilisation of Structure Based Design to Identify Novel, Irreversible Inhibitors of the Epidermal Growth Factor Receptor (Egfr) Harboring the Gatekeeper T790M Mutation
To be Published
|
|
3D3Q
 
 | | Crystal structure of tRNA delta(2)-isopentenylpyrophosphate transferase (SE0981) from Staphylococcus epidermidis. Northeast Structural Genomics Consortium target SeR100 | | Descriptor: | tRNA delta(2)-isopentenylpyrophosphate transferase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, L, Xiao, R, Maglaqui, M, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of tRNA delta(2)-isopentenylpyrophosphate transferase (SE0981) from Staphylococcus epidermidis.
To be Published
|
|
3CEI
 
 | | Crystal Structure of Superoxide Dismutase from Helicobacter pylori | | Descriptor: | FE (III) ION, SULFATE ION, Superoxide dismutase | | Authors: | Esposito, L, Seydel, A, Aiello, R, Sorrentino, G, Cendron, L, Zanotti, G, Zagari, A. | | Deposit date: | 2008-02-29 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the superoxide dismutase from Helicobacter pylori reveals a structured C-terminal extension
Biochim.Biophys.Acta, 1784, 2008
|
|
1BK6
 
 | | KARYOPHERIN ALPHA (YEAST) + SV40 T ANTIGEN NLS | | Descriptor: | KARYOPHERIN ALPHA, LARGE T ANTIGEN | | Authors: | Conti, E, Uy, M, Leighton, L, Blobel, G, Kuriyan, J. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BK5
 
 | | KARYOPHERIN ALPHA FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | COBALT (II) ION, KARYOPHERIN ALPHA | | Authors: | Conti, E, Uy, M, Leighton, L, Blobel, G, Kuriyan, J. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BOX
 
 | | N39S MUTANT OF RNASE SA FROM STREPTOMYCES AUREOFACIENS | | Descriptor: | GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Hebert, E.J, Giletto, A, Sevcik, J, Urbanikova, L, Wilson, K.S, Dauter, Z, Pace, C.N. | | Deposit date: | 1998-08-07 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Contribution of a conserved asparagine to the conformational stability of ribonucleases Sa, Ba, and T1.
Biochemistry, 37, 1998
|
|
1BN6
 
 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
1BS0
 
 | | PLP-DEPENDENT ACYL-COA SYNTHASE | | Descriptor: | PROTEIN (8-AMINO-7-OXONANOATE SYNTHASE), SULFATE ION | | Authors: | Alexeev, D, Alexeeva, M, Baxter, R.L, Campopiano, D.J, Webster, S.P, Sawyer, L. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of 8-amino-7-oxononanoate synthase: a bacterial PLP-dependent, acyl-CoA-condensing enzyme.
J.Mol.Biol., 284, 1998
|
|
1BXD
 
 | | NMR STRUCTURE OF THE HISTIDINE KINASE DOMAIN OF THE E. COLI OSMOSENSOR ENVZ | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (OSMOLARITY SENSOR PROTEIN (ENVZ)) | | Authors: | Tanaka, T, Saha, S.K, Tomomori, C, Ishima, R, Liu, D, Tong, K.I, Park, H, Dutta, R, Qin, L, Swindells, M.B, Yamazaki, T, Ono, A.M, Kainosho, M, Inouye, M, Ikura, M. | | Deposit date: | 1998-10-02 | | Release date: | 1999-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ.
Nature, 396, 1998
|
|
1BYX
 
 | | CHIMERIC HYBRID DUPLEX R(GCAGUGGC).R(GCCA)D(CTGC) COMPRISING THE TRNA-DNA JUNCTION FORMED DURING INITIATION OF HIV-1 REVERSE TRANSCRIPTION | | Descriptor: | DNA/RNA (5'-R(*GP*CP*CP*A)-D(P*CP*TP*GP*C)-3'), RNA (5'-R(*GP*CP*AP*GP*UP*GP*GP*C)-3') | | Authors: | Szyperski, T, Goette, M, Billeter, M, Perola, E, Cellai, L. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the chimeric hybrid duplex r(gcaguggc).r(gcca)d(CTGC) comprising the tRNA-DNA junction formed during initiation of HIV-1 reverse transcription.
J.Biomol.NMR, 13, 1999
|
|
1BJJ
 
 | | AGKISTRODOTOXIN, A PHOSPHOLIPASE A2-TYPE PRESYNAPTIC NEUROTOXIN FROM AGKISTRODON HALYS PALLAS | | Descriptor: | AGKISTRODOTOXIN, CALCIUM ION | | Authors: | Tang, L, Zhou, Y, Lin, Z. | | Deposit date: | 1998-06-25 | | Release date: | 1999-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of agkistrodotoxin in an orthorhombic crystal form with six molecules per asymmetric unit.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BY5
 
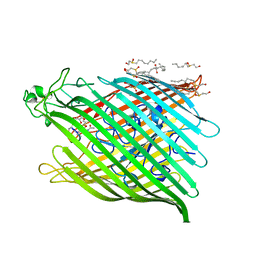 | | FHUA FROM E. COLI, WITH ITS LIGAND FERRICHROME | | Descriptor: | FE (III) ION, FERRIC HYDROXAMATE UPTAKE PROTEIN, FERRICHROME, ... | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-23 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
1C4O
 
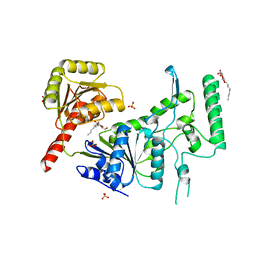 | | CRYSTAL STRUCTURE OF THE DNA NUCLEOTIDE EXCISION REPAIR ENZYME UVRB FROM THERMUS THERMOPHILUS | | Descriptor: | DNA NUCLEOTIDE EXCISION REPAIR ENZYME UVRB, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Machius, M, Henry, L, Palnitkar, M, Deisenhofer, J. | | Deposit date: | 1999-09-14 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
