3W40
 
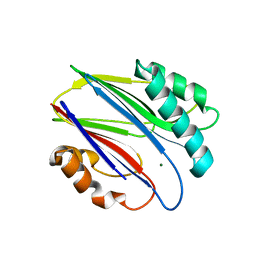 | | Crystal structure of RsbX in complex with magnesium in space group P1 | | Descriptor: | MAGNESIUM ION, Phosphoserine phosphatase RsbX | | Authors: | Teh, A.H, Makino, M, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W43
 
 | | Crystal structure of RsbX in complex with manganese in space group P21 | | Descriptor: | MANGANESE (II) ION, Phosphoserine phosphatase RsbX | | Authors: | Teh, A.H, Makino, M, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W45
 
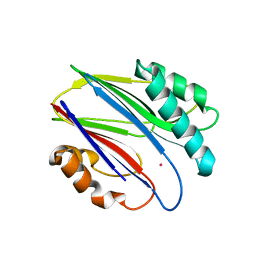 | | Crystal structure of RsbX in complex with cobalt in space group P1 | | Descriptor: | COBALT (II) ION, Phosphoserine phosphatase RsbX | | Authors: | Makino, M, Teh, A.H, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1WQS
 
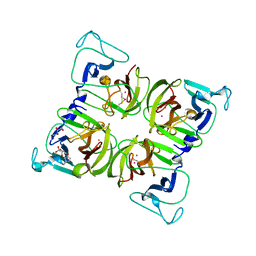 | | Crystal structure of Norovirus 3C-like protease | | Descriptor: | 3C-like protease, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Nakamura, K, Someya, Y, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-10-01 | | Release date: | 2005-10-04 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A norovirus protease structure provides insights into active and substrate binding site integrity
J.Virol., 79, 2005
|
|
6J8M
 
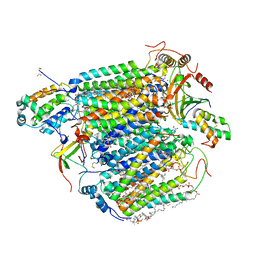 | | Low-dose structure of bovine heart cytochrome c oxidase in the fully oxidized state determined using 30 keV X-ray | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ueno, G, Shimada, A, Yamashita, E, Hasegawa, K, Kumasaka, T, Shinzawa-Itoh, K, Yoshikawa, S, Tsukihara, T, Yamamoto, M. | | Deposit date: | 2019-01-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Low-dose X-ray structure analysis of cytochrome c oxidase utilizing high-energy X-rays.
J.Synchrotron Radiat., 26, 2019
|
|
1WPR
 
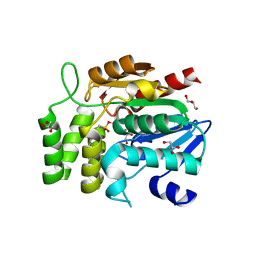 | | Crystal structure of RsbQ inhibited by PMSF | | Descriptor: | GLYCEROL, Sigma factor sigB regulation protein rsbQ, phenylmethanesulfonic acid | | Authors: | Kaneko, T, Tanaka, N, Kumasaka, T. | | Deposit date: | 2004-09-11 | | Release date: | 2005-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
1WOM
 
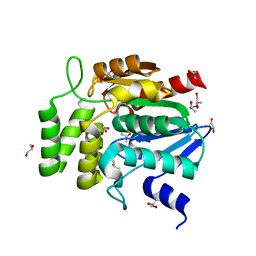 | | Crystal structure of RsbQ | | Descriptor: | MALONIC ACID, S-1,2-PROPANEDIOL, Sigma factor sigB regulation protein rsbQ | | Authors: | Kaneko, T, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-08-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
3ANX
 
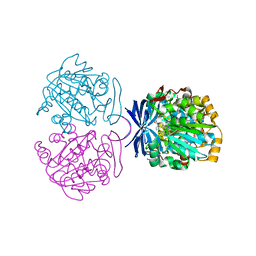 | | Crystal structure of triamine/agmatine aminopropyltransferase (SPEE) from thermus thermophilus, complexed with MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, spermidine synthase | | Authors: | Ganbe, T, Ohnuma, M, Sato, T, Tanaka, N, Oshima, T, Kumasaka, T. | | Deposit date: | 2010-09-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures and enzymatic properties of a triamine/agmatine aminopropyltransferase from Thermus thermophilus
J.Mol.Biol., 408, 2011
|
|
2ZTW
 
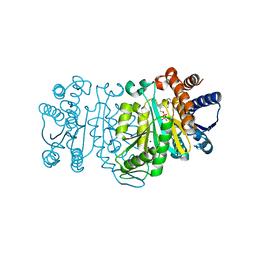 | | Structure of 3-isopropylmalate dehydrogenase in complex with the inhibitor and NAD+ | | Descriptor: | (2Z)-2-hydroxy-3-(methylsulfanyl)prop-2-enoic acid, 3-isopropylmalate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Nango, E, Kumasaka, T, Eguchi, T. | | Deposit date: | 2008-10-10 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of 3-isopropylmalate dehydrogenase in complex with NAD(+) and a designed inhibitor
Bioorg.Med.Chem., 17, 2009
|
|
6K55
 
 | | Inactivated mutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide | | Descriptor: | Glucanase, MAGNESIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
6K54
 
 | | Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with trisaccharide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Novel hyperthermophilic cellobiohydrolase II isolated from hot spring microbial community
To Be Published
|
|
6K53
 
 | | A variant of metagenome-derived GH6 cellobiohydrolase, HmCel6A (P88S/L230F/F414S) | | Descriptor: | CITRATE ANION, GH6 cellobiohydrolase, HMCEL6A, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A hyperthermophilic GH6 cellobiohydrolase (HmCel6A) from a hot spring metagenomic data
To Be Published
|
|
6K52
 
 | | Hyperthermophilic GH6 cellobiohydrolase (HmCel6A) from the microbial flora of a Japanese hot spring | | Descriptor: | ACETATE ION, CALCIUM ION, GH6 cellobiohydrolase, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68000138 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
3AGG
 
 | | X-ray analysis of lysozyme in the absence of Arg | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
3AGH
 
 | | X-ray analysis of lysozyme in the presence of 200 mM Arg | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
3AGI
 
 | | High resolution X-ray analysis of Arg-lysozyme complex in the presence of 500 mM Arg | | Descriptor: | ACETATE ION, ARGININE, CHLORIDE ION, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
1K1X
 
 | | Crystal structure of 4-alpha-glucanotransferase from thermococcus litoralis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
1K1Y
 
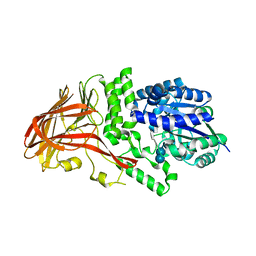 | | Crystal structure of thermococcus litoralis 4-alpha-glucanotransferase complexed with acarbose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-ALPHA-GLUCANOTRANSFERASE, ... | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
1K1W
 
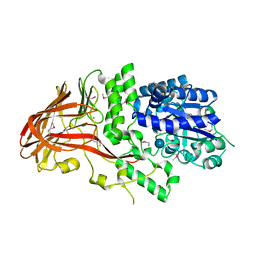 | | Crystal structure of 4-alpha-glucanotransferase from thermococcus litoralis | | Descriptor: | 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
3AJN
 
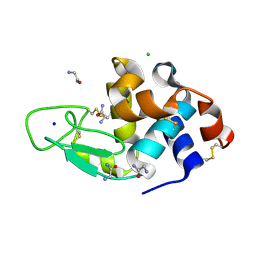 | | Structural basis of glycine amide on suppression of protein aggregation by high resolution X-ray analysis | | Descriptor: | AMINOMETHYLAMIDE, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Kumasaka, T. | | Deposit date: | 2010-06-09 | | Release date: | 2011-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Glycine amide shielding on the aromatic surfaces of lysozyme: Implication for suppression of protein aggregation
Febs Lett., 585, 2011
|
|
2ZW2
 
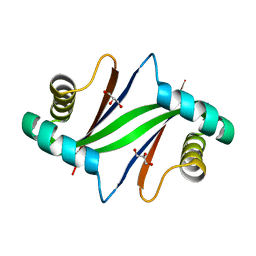 | | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS) | | Descriptor: | GLYCEROL, Putative uncharacterized protein STS178 | | Authors: | Suzuki, S, Tamura, S, Okada, K, Baba, S, Kumasaka, T, Nakagawa, N, Masui, R, Kuramitsu, S, Sampei, G, Kawai, G. | | Deposit date: | 2008-11-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS)
To be Published
|
|
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X1F
 
 | | CO bound cytochrome c oxidase without pump laser irradiation at 278K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
1IO1
 
 | | CRYSTAL STRUCTURE OF F41 FRAGMENT OF FLAGELLIN | | Descriptor: | PHASE 1 FLAGELLIN | | Authors: | Samatey, F.A, Imada, K, Nagashima, S, Vondervisz, F, Kumasaka, T, Yamamoto, M, Namba, K. | | Deposit date: | 2000-12-28 | | Release date: | 2001-04-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling
Nature, 410, 2001
|
|
