4JNU
 
 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group P21 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JQ5
 
 | |
4JO9
 
 | | Crystal structure of the human Nup49CCS2+3* Nup57CCS3* complex 1:2 stoichiometry | | Descriptor: | Nucleoporin p54, Nucleoporin p58/p45 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JO7
 
 | | Crystal structure of the human Nup49CCS2+3* Nup57CCS3* complex with 2:2 stoichiometry | | Descriptor: | Nucleoporin p54, Nucleoporin p58/p45 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-17 | | Release date: | 2014-09-17 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JNV
 
 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group C2 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
4JE4
 
 | | Crystal Structure of Monobody NSa1/SHP2 N-SH2 Domain Complex | | Descriptor: | Monobody NSa1, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sha, F, Koide, S. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JMG
 
 | |
4JEG
 
 | | Crystal Structure of Monobody CS1/SHP2 C-SH2 Domain Complex | | Descriptor: | Monobody CS1, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sha, F, Koide, S. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-28 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JMH
 
 | |
7MH6
 
 | |
7MGX
 
 | |
1X0C
 
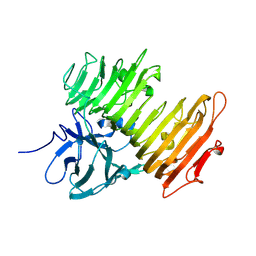 | | Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase | | Authors: | Mizuno, M, Tonozuka, T, Yamamura, A, Miyasaka, Y, Akeboshi, H, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-17 | | Release date: | 2006-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
6UX8
 
 | |
1WMR
 
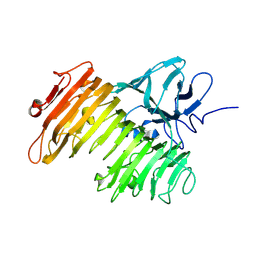 | | Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase | | Authors: | Mizuno, M, Tonozuka, T, Miyasaka, Y, Akeboshi, H, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
6WU1
 
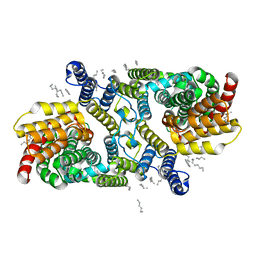 | | Structure of apo LaINDY | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N.C, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WTX
 
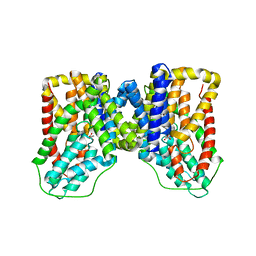 | |
6WK5
 
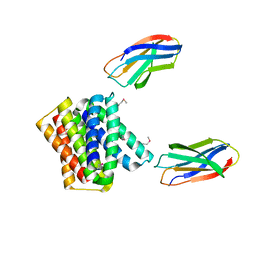 | |
6WK9
 
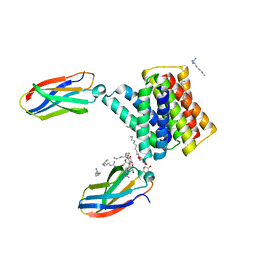 | |
6WU4
 
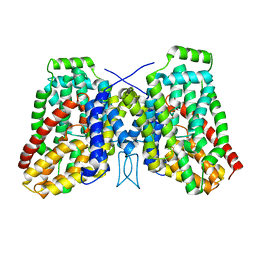 | | Structure of the LaINDY-alpha-ketoglutarate complex | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU3
 
 | |
6WU2
 
 | | Structure of the LaINDY-malate complex | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WK8
 
 | |
6WTW
 
 | | Structure of LaINDY crystallized in the presence of alpha-ketoglutarate and malate | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Cocco, N, Marden, J.J, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6CPG
 
 | | Structure of dephosphorylated Aurora A (122-403) in complex with inhibiting monobody and AT9283 in an inactive conformation | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Aurora kinase A, Monobody | | Authors: | Otten, R, Kutter, S, Zorba, A, Padua, R.A.P, Koide, A, Koide, S, Kern, D. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dynamics of human protein kinase Aurora A linked to drug selectivity.
Elife, 7, 2018
|
|
2OYB
 
 | |
