5GGO
 
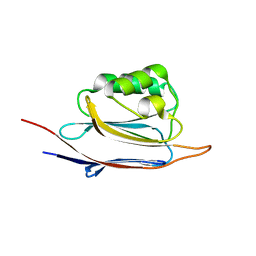 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GalNac-beta1,3-GlcNAc-beta-pNP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8XBI
 
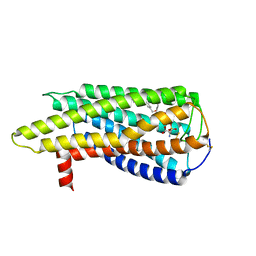 | | Human GPR34 -Gi complex bound to M1, receptor focused | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Probable G-protein coupled receptor 34 | | Authors: | Kawahara, R, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XBH
 
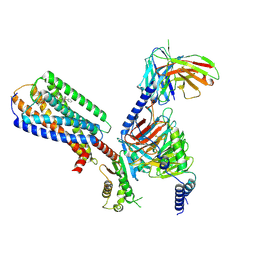 | | Human GPR34 -Gi complex bound to M1 | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Kawahara, R, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XBG
 
 | |
8XBE
 
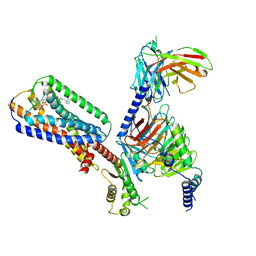 | | Human GPR34 -Gi complex bound to S3E-LysoPS | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[(~{Z})-octadec-9-enoyl]oxy-propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Kawahara, R, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-06 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
5GGF
 
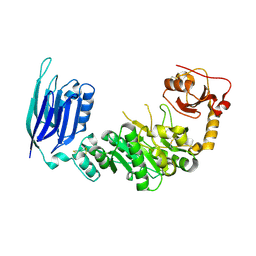 | |
5GGJ
 
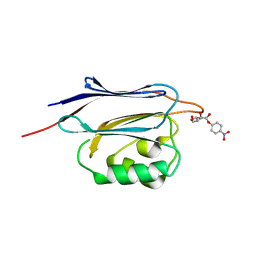 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Man-alpha-pNP | | Descriptor: | 4-nitrophenyl alpha-D-mannopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3RP2
 
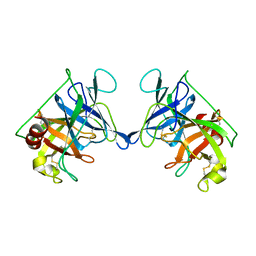 | | THE STRUCTURE OF RAT MAST CELL PROTEASE II AT 1.9-ANGSTROMS RESOLUTION | | Descriptor: | RAT MAST CELL PROTEASE II | | Authors: | Reynolds, R, Remington, S, Weaver, L, Fischer, R, Anderson, W, Ammon, H, Matthews, B. | | Deposit date: | 1984-09-10 | | Release date: | 1984-10-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of rat mast cell protease II at 1.9-A resolution.
Biochemistry, 27, 1988
|
|
7CTQ
 
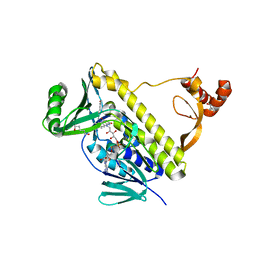 | | Peptidyl tryptophan dihydroxylase QhpG essential for tryptophylquinone cofactor biogenesis | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S},4~{R},5~{R},6~{R})-6-(cyclohexylmethoxy)-2-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-3-yl]oxy-6-(hydroxymethyl)oxane-3,4,5-triol, FLAVIN-ADENINE DINUCLEOTIDE, HEXANE-1,6-DIOL, ... | | Authors: | Oozeki, T, Nakai, T, Okajima, T. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Functional and structural characterization of a flavoprotein monooxygenase essential for biogenesis of tryptophylquinone cofactor.
Nat Commun, 12, 2021
|
|
7VR7
 
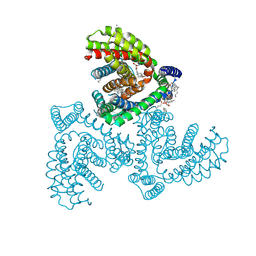 | | Inward-facing structure of human EAAT2 in the WAY213613-bound state | | Descriptor: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
7E18
 
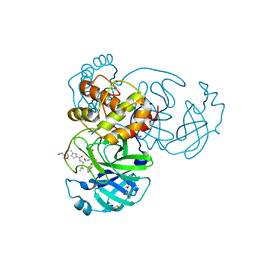 | | Crystal structure of SAR-CoV-2 3CL protease complex with inhibitor YH-53 | | Descriptor: | 1,2-ETHANEDIOL, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1ab | | Authors: | Senda, M, Konno, S, Hayashi, Y, Senda, T. | | Deposit date: | 2021-02-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents.
J.Med.Chem., 65, 2022
|
|
7E19
 
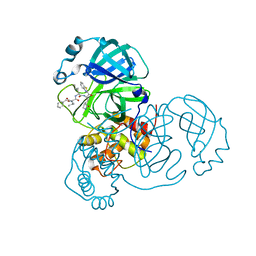 | | Crystal structure of SAR-CoV-2 3CL protease complex with inhibitor SH-5 | | Descriptor: | (phenylmethyl) N-[(2S)-1-[[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]carbamate, 3C-like proteinase | | Authors: | Senda, M, Konno, S, Hayashi, Y, Senda, T. | | Deposit date: | 2021-02-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents.
J.Med.Chem., 65, 2022
|
|
7VR8
 
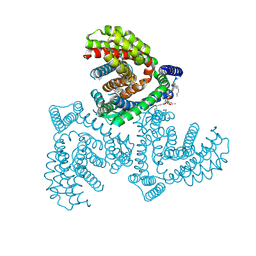 | | Inward-facing structure of human EAAT2 in the substrate-free state | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
7XJI
 
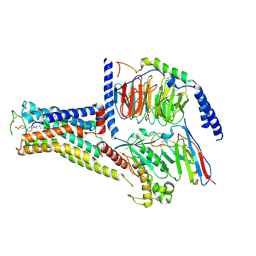 | | Solabegron-activated dog beta3 adrenergic receptor | | Descriptor: | 3-[3-[2-[[(2~{S})-2-(3-chlorophenyl)-2-oxidanyl-ethyl]amino]ethylamino]phenyl]benzoic acid, Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the beta 3 adrenergic receptor bound to solabegron and isoproterenol.
Biochem.Biophys.Res.Commun., 611, 2022
|
|
2ZOO
 
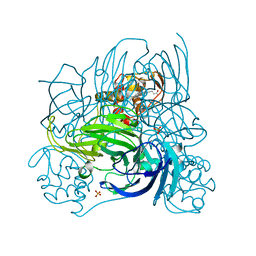 | | Crystal structure of nitrite reductase from Pseudoalteromonas haloplanktis TAC125 | | Descriptor: | COPPER (II) ION, PROTOPORPHYRIN IX CONTAINING FE, Probable nitrite reductase, ... | | Authors: | Nojiri, M, Tsuda, A, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2008-05-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Electron transfer processes within and between proteins containing the HEME C and blue Cu
To be Published
|
|
2ZON
 
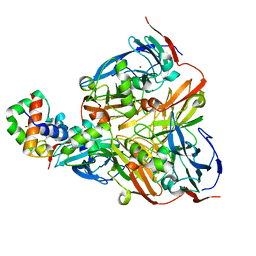 | | Crystal structure of electron transfer complex of nitrite reductase with cytochrome c | | Descriptor: | COPPER (II) ION, Dissimilatory copper-containing nitrite reductase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Nojiri, M, Koteishi, H, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2008-05-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of inter-protein electron transfer for nitrite reduction in denitrification
Nature, 462, 2009
|
|
7E7Z
 
 | |
7E83
 
 | |
7E87
 
 | |
7E8H
 
 | | CryoEM structure of human Kv4.2-DPP6S-KChIP1 complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7E84
 
 | | CryoEM structure of human Kv4.2-KChIP1 complex | | Descriptor: | Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-02-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7E8E
 
 | |
7E8G
 
 | |
7E89
 
 | |
7E8B
 
 | | CryoEM structure of human Kv4.2-DPP6S complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
