7TYZ
 
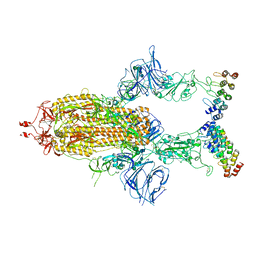 | | Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DARPin FSR22, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
5UG3
 
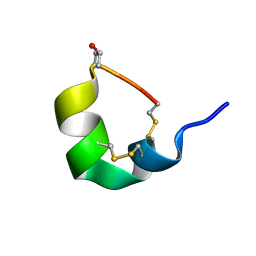 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID MUTANT A10V | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A.K, Leffler, A.E, Zebroski, H.A, Powell, S.R, Kuryatov, A, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UG5
 
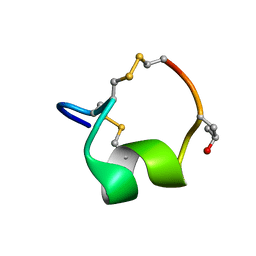 | | NMR SOLUTION STRUCTURE OF THE ALPHA-CONOTOXIN GID MUTANT V13Y | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A, Leffler, A.E, Kuryatov, A, Zebroski, H.A, Powell, S.R, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1XXL
 
 | |
1Y7E
 
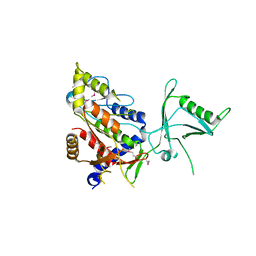 | |
1Y9E
 
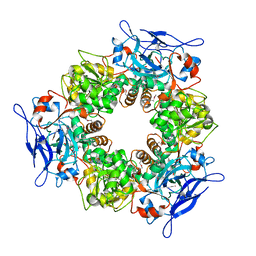 | |
8DW2
 
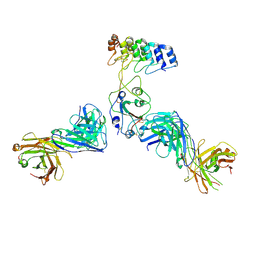 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Antibody CR3022 heavy chain, Antibody CR3022 light chain, Antibody S309 heavy chain, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8DW3
 
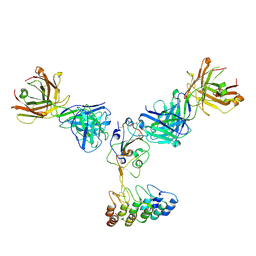 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Anti-SARS-CoV-2 DARPin SR16m, Antibody S309 light chain, Spike protein S1, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
2IXP
 
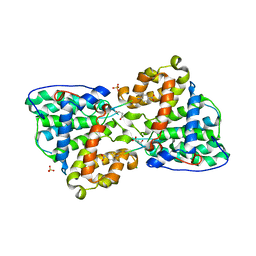 | | Crystal structure of the Pp2A phosphatase activator Ypa1 PTPA1 in complex with model substrate | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN PHOSPHATASE 2A ACTIVATOR 1, SIN-ALA-ALA-PRO-LYS-NIT, ... | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the PP2A phosphatase activator: implications for its PP2A-specific PPIase activity.
Mol. Cell, 23, 2006
|
|
2IXN
 
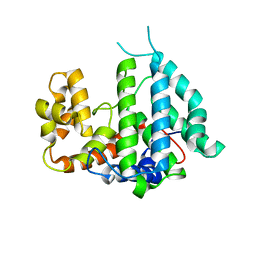 | | CRYSTAL STRUCTURE OF THE PP2A PHOSPHATASE ACTIVATOR Ypa2 PTPA2 | | Descriptor: | SERINE/THREONINE-PROTEIN PHOSPHATASE 2A ACTIVATOR 2 | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Pp2A Phosphatase Activator: Implications for its Pp2A-Specific Ppiase Activity
Mol.Cell, 23, 2006
|
|
2IXM
 
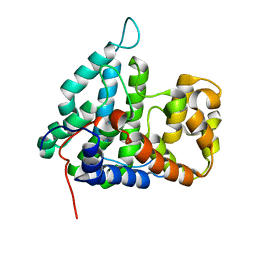 | | Structure of human PTPA | | Descriptor: | SERINE/THREONINE-PROTEIN PHOSPHATASE 2A REGULATORY SUBUNIT B' | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the Pp2A Phosphatase Activator: Implications for its Pp2A-Specific Ppiase Activity
Mol.Cell, 23, 2006
|
|
2IXO
 
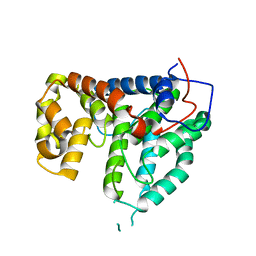 | | CRYSTAL STRUCTURE OF THE PP2A PHOSPHATASE ACTIVATOR Ypa1 PTPA1 | | Descriptor: | SERINE/THREONINE-PROTEIN PHOSPHATASE 2A ACTIVATOR 1 | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Pp2A Phosphatase Activator: Implications for its Pp2A-Specific Ppiase Activity.
Mol.Cell, 23, 2006
|
|
1ZSJ
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in complex with N-(7-Carbamimidoyl-naphthalen-1-yl)-3-hydroxy-2-methyl-benzamide | | Descriptor: | BICARBONATE ION, Coagulation factor XI, N-(7-CARBAMIMIDOYL-NAPHTHALEN-1-YL)-3-HYDROXY-2-METHYL-BENZAMIDE, ... | | Authors: | Guo, Z, Bannister, T, Noll, R, Jin, L, Rynkiewicz, M, Bibbins, F, Magee, S, Gorga, J, Babine, R.E, Strickler, J.E, Meyers, H.V, Abdel-Meguid, S.S. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and Optimization of Potent and Selective Inhibitors for Human Factor XIa: Substituted Naphthamidine Series
To be Published
|
|
1ZSK
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 6-Carbamimidoyl-4-(3-hydroxy-2-methyl-benzoylamino)-naphthalene-2-carboxylic acid methyl ester | | Descriptor: | 6-CARBAMIMIDOYL-4-(3-HYDROXY-2-METHYL-BENZOYLAMINO)-NAPHTHALENE-2-CARBOXYLIC ACID METHYL ESTER, BICARBONATE ION, Coagulation factor XI, ... | | Authors: | Guo, Z, Bannister, T, Noll, R, Jin, L, Rynkiewicz, M, Bibbins, F, Magee, S, Gorga, J, Babine, R.E, Strickler, J.E, Meyers, H.V, Abdel-Meguid, S.S. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and Optimization of Potent and Selective Inhibitors for Human Factor XIa: Substituted Naphthamidine Series
To be Published
|
|
2GLU
 
 | | The crystal structure of YcgJ protein from Bacillus subitilis | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, ycgJ | | Authors: | Burke, T, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The crystal structure of YcgJ protein from Bacillus subitilis
To be Published
|
|
1Q7E
 
 | | Crystal Structure of YfdW protein from E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hypothetical protein yfdW, METHIONINE | | Authors: | Gogos, A, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-18 | | Release date: | 2003-09-30 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Escherichia coli YfdW, a type III CoA transferase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1Q6Y
 
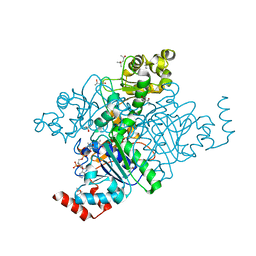 | | Hypothetical protein YfdW from E. coli bound to Coenzyme A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COENZYME A, Hypothetical protein yfdW | | Authors: | Gogos, A, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-14 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of Escherichia coli YfdW, a type III CoA transferase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PQY
 
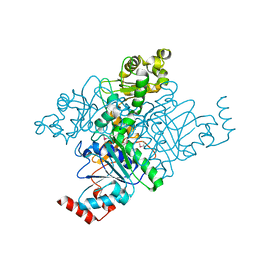 | | Crystal structure of formyl-coA transferase yfdW from E. coli | | Descriptor: | Hypothetical protein yfdW | | Authors: | Gogos, A, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-19 | | Release date: | 2003-09-30 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Escherichia coli YfdW, a type III CoA transferase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T5X
 
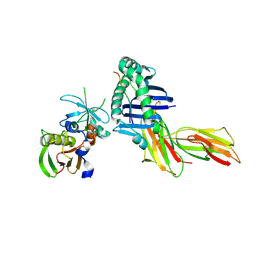 | | HLA-DR1 in complex with a synthetic peptide (AAYSDQATPLLLSPR) and the superantigen SEC3-3B2 | | Descriptor: | 15-mer peptide fragment of Regulatory protein MIG1, Enterotoxin type C-3, HLA class II histocompatibility antigen, ... | | Authors: | Zavala-Ruiz, Z, Strug, I, Anderson, M.W, Gorski, J, Stern, L.J. | | Deposit date: | 2004-05-05 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Polymorphic Pocket at the P10 Position Contributes to Peptide Binding Specificity in Class II MHC Proteins
Chem.Biol., 11, 2004
|
|
2NYV
 
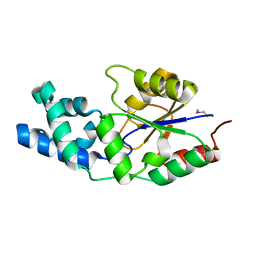 | | X-ray crystal structure of a phosphoglycolate phosphatase from Aquifex aeolicus | | Descriptor: | Phosphoglycolate phosphatase | | Authors: | Ciatto, C, Min, T, Gorman, J, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | X-ray crystal structure of a phosphoglycolate phosphatase from Aquifex aeolicus
To be Published
|
|
1T5W
 
 | | HLA-DR1 in complex with a synthetic peptide (AAYSDQATPLLLSPR) | | Descriptor: | 15-mer peptide fragment of Regulatory protein MIG1, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Zavala-Ruiz, Z, Strug, I, Anderson, M.W, Gorski, J, Stern, L.J. | | Deposit date: | 2004-05-05 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Polymorphic Pocket at the P10 Position Contributes to Peptide Binding Specificity in Class II MHC Proteins
Chem.Biol., 11, 2004
|
|
1T5J
 
 | |
1TT7
 
 | |
1TSJ
 
 | |
5T33
 
 | | Crystal structure of strain-specific glycan-dependent CD4 binding site-directed neutralizing antibody CAP257-RH1, in complex with HIV-1 strain RHPA gp120 core with an oligomannose N276 glycan. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CAP257-RH1 heavy chain, ... | | Authors: | Wibmer, C.K, Gorman, J, Kwong, P.D. | | Deposit date: | 2016-08-24 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2092 Å) | | Cite: | Structure of an N276-Dependent HIV-1 Neutralizing Antibody Targeting a Rare V5 Glycan Hole Adjacent to the CD4 Binding Site.
J.Virol., 90, 2016
|
|
