8WQD
 
 | |
8WQC
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CDK5R1 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQG
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
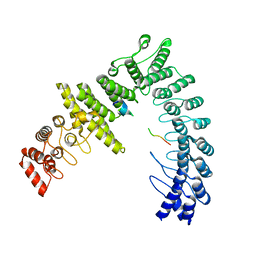
8WQI
 
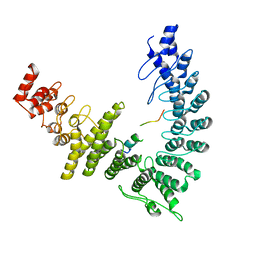 | |
8WQB
 
 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
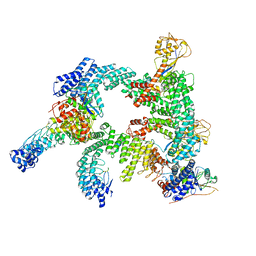
8WQA
 
 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQH
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8GI6
 
 | | Crystal structure of RhoA mutant L69R complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ras-like GTPases mutant structures
To be published
|
|
8GI3
 
 | | Crystal structure of RhoA mutant L69P complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ras-like GTPases mutants structure
To be published
|
|
6MUN
 
 | | Structure of hRpn10 bound to UBQLN2 UBL | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquilin-2 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2018-10-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome.
J.Mol.Biol., 431, 2019
|
|
5IRS
 
 | | crystal structure of the proteasomal Rpn13 PRU-domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Proteasomal ubiquitin receptor ADRM1 | | Authors: | Chen, X, Shi, K, Walters, K, Aihara, H. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
6OEO
 
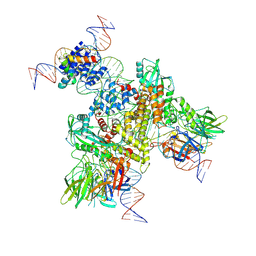 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEQ
 
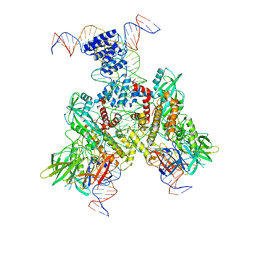 | | Cryo-EM structure of mouse RAG1/2 12RSS-PRC/23RSS-NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OER
 
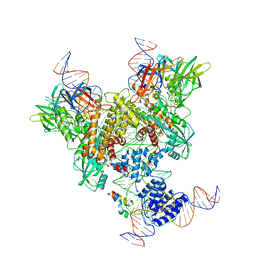 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA2) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6K9C
 
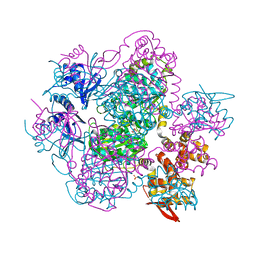 | | The apo structure of NrS-1 C terminal region (305-718) | | Descriptor: | MERCURY (II) ION, Primase, SULFATE ION | | Authors: | Chen, X, Gan, J. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structural studies reveal a ring-shaped architecture of deep-sea vent phage NrS-1 polymerase.
Nucleic Acids Res., 48, 2020
|
|
6K9E
 
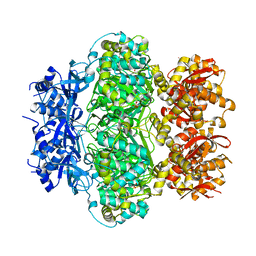 | |
8WQ3
 
 | | Crystal structure of the C-terminal RRM domain of an RBP | | Descriptor: | CHLORIDE ION, RNA-binding protein 45 | | Authors: | Chen, X, Jiang, M, Yang, Z, Chen, X, Wei, Q, Guo, S, Wang, M. | | Deposit date: | 2023-10-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for RNA recognition by the C-terminal RRM domain of human RBM45.
J.Biol.Chem., 300, 2024
|
|
8WQ5
 
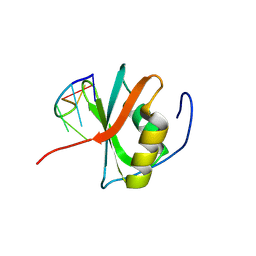 | | Crystal structure of the C-terminal RRM domain of an RBP in complex with ssDNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*GP*CP*AP*G)-3'), GLYCEROL, RNA-binding protein 45 | | Authors: | Chen, X, Yang, Z, Chen, X, Wei, Q, Guo, S, Jiang, M, Wang, M. | | Deposit date: | 2023-10-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for RNA recognition by the C-terminal RRM domain of human RBM45.
J.Biol.Chem., 300, 2024
|
|
8U5H
 
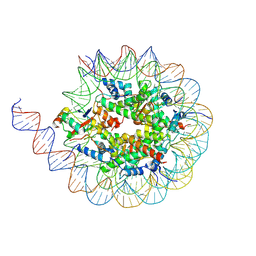 | |
6LBG
 
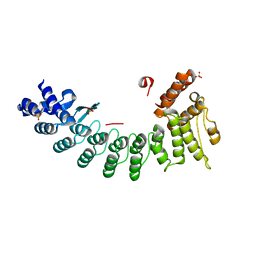 | | Structure of OR51B2 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Olfactory receptor 51B2, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LE6
 
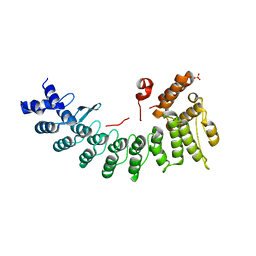 | | Structure of LNLPTQGRAR bound FEM1C | | Descriptor: | Protein fem-1 homolog C,10-mer peptide, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-24 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
5CX3
 
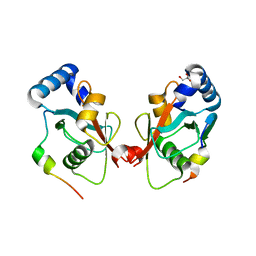 | | Crystal structure of FYCO1 LIR in complex with LC3A | | Descriptor: | FYVE and coiled-coil domain-containing protein 1, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Cheng, X, Pan, L. | | Deposit date: | 2015-07-28 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of FYCO1 and MAP1LC3A interaction reveals a novel binding mode for Atg8-family proteins.
Autophagy, 12, 2016
|
|
7DWS
 
 | |
8GRD
 
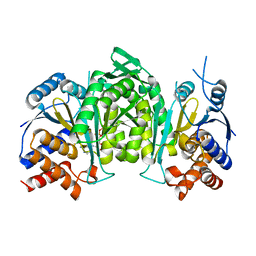 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ADP and Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, ... | | Authors: | Chen, X, Sun, P, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8XKF
 
 | | Crystal structure of Helicobacter pylori IspDF with substrate CTP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
