4GEG
 
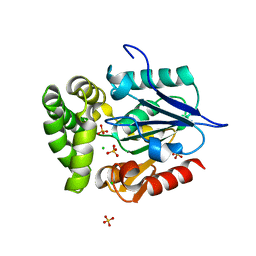 | | Crystal Structure of E.coli MenH Y85F Mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, CHLORIDE ION, ... | | Authors: | Johnston, J.M, Baker, E.N, Guo, Z, Jiang, M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal Structures of E. coli Native MenH and Two Active Site Mutants.
Plos One, 8, 2013
|
|
4GIU
 
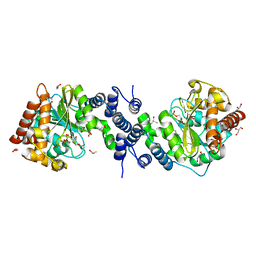 | | Bianthranilate-like analogue bound in inner site of anthranilate phosphoribosyltransferase (AnPRT; trpD). | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-[(2-carboxy-5-methylphenyl)amino]-3-methylbenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Evans, G.L, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-08-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | Repurposing the Chemical Scaffold of the Anti-Arthritic Drug Lobenzarit to Target Tryptophan Biosynthesis in Mycobacterium tuberculosis.
Chembiochem, 15, 2014
|
|
4HSS
 
 | |
4HSQ
 
 | |
5C7S
 
 | |
5BYT
 
 | |
5CEV
 
 | | ARGINASE FROM BACILLUS CALDEVELOX, L-LYSINE COMPLEX | | Descriptor: | GUANIDINE, LYSINE, MANGANESE (II) ION, ... | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-16 | | Release date: | 1999-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
5E5Q
 
 | | Racemic snakin-1 in P21/c | | Descriptor: | Snakin-1 | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5Y
 
 | | Quasi-racemic snakin-1 in P1 before radiation damage | | Descriptor: | 1,2-ETHANEDIOL, D- snakin-1, FORMIC ACID, ... | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5T
 
 | | Quasi-racemic snakin-1 in P1 after radiation damage | | Descriptor: | 1,2-ETHANEDIOL, D- snakin-1, FORMIC ACID, ... | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5CXI
 
 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with 3-oxo-23,24-bisnorchol-4-en-22-oyl-CoA (4-BNC-CoA) | | Descriptor: | 3-oxo-23,24-bisnorchol-4-en-22-oyl-CoA, HTH-type transcriptional repressor KstR | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Casabon, I, Crowe, A.M, Baker, E.N, Eltis, L.D, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
1LCF
 
 | | CRYSTAL STRUCTURE OF COPPER-AND OXALATE-SUBSTITUTED HUMAN LACTOFERRIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, COPPER (II) ION, ... | | Authors: | Smith, C.A, Anderson, B.F, Baker, H.M, Baker, E.N. | | Deposit date: | 1994-01-11 | | Release date: | 1994-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of copper- and oxalate-substituted human lactoferrin at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1LFI
 
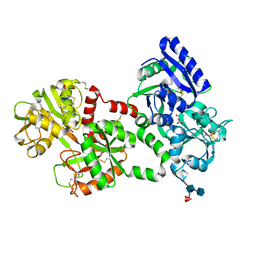 | | METAL SUBSTITUTION IN TRANSFERRINS: THE CRYSTAL STRUCTURE OF HUMAN COPPER-LACTOFERRIN AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, COPPER (II) ION, ... | | Authors: | Smith, C.A, Anderson, B.F, Baker, H.M, Baker, E.N. | | Deposit date: | 1992-02-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metal substitution in transferrins: the crystal structure of human copper-lactoferrin at 2.1-A resolution.
Biochemistry, 31, 1992
|
|
5CW8
 
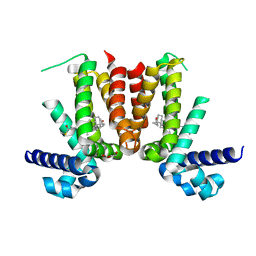 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with 3-oxo-4-cholesten-26-oyl-CoA | | Descriptor: | HTH-type transcriptional repressor KstR, S-{1-[5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,7-dihydroxy-6,6-dimethyl-3-oxido-8,12 -dioxo-2,4-dioxa-9,13-diaza-3lambda~5~-phosphapentadecan-15-yl} (2S,6R)-6-[(8S,9S,10R,13R,14S,17R)-10,13-dimethyl-3-oxo-2,3,6,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta [a]phenanthren-17-yl]-2-methylheptanethioate (non-preferred name), TRIETHYLENE GLYCOL | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Casabon, I, Crowe, A.M, Baker, E.N, Eltis, L.D, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-27 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5CXG
 
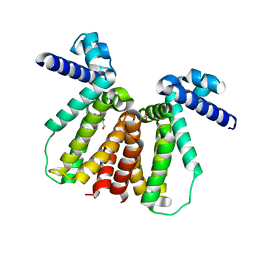 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with PEG | | Descriptor: | DI(HYDROXYETHYL)ETHER, HTH-type transcriptional repressor KstR, TRIETHYLENE GLYCOL | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1001 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5CYY
 
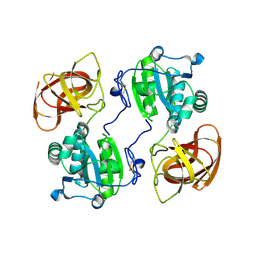 | |
5C1R
 
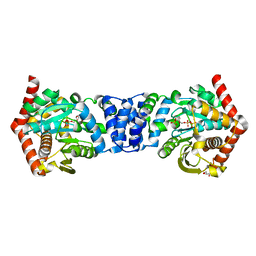 | | Stereoisomer of PRPP bound in the active site of Mycobacterium tuberculosis anthranilate phosphoribosyl (AnPRT; trpD) | | Descriptor: | 5-O-[(R)-hydroxy(phosphonooxy)phosphoryl]-1-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, GLYCEROL, ... | | Authors: | Evans, G.L, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-06-15 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Binding and mimicking of the phosphate-rich substrate, PRPP.
To Be Published
|
|
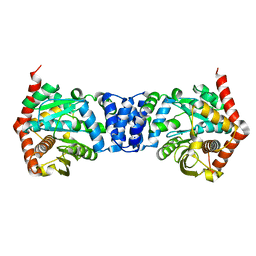
5C2L
 
 | |
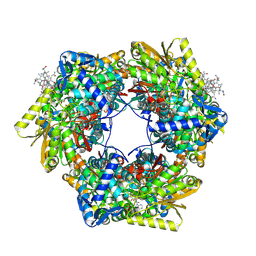
4NS3
 
 | |
4P0D
 
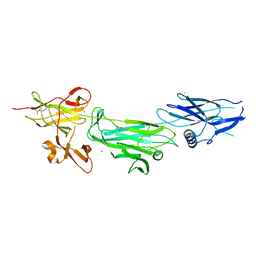 | | The T6 backbone pilin of serotype M6 Streptococcus pyogenes has a modular three-domain structure decorated with variable loops and extensions | | Descriptor: | CALCIUM ION, IODIDE ION, Trypsin-resistant surface T6 protein | | Authors: | Young, P.G, Moreland, N.J, Loh, J.M, Bell, A, Atatoa-Carr, P, Proft, T, Baker, E.N. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Conservation, Variability, and Immunogenicity of the T6 Backbone Pilin of Serotype M6 Streptococcus pyogenes.
Infect.Immun., 82, 2014
|
|
4PMK
 
 | |
4N8Q
 
 | | Alternative substrates of Mycobacterium tuberculosis anthranilate phosphoribosyl transferase | | Descriptor: | 2-amino-4-fluorobenzoic acid, Anthranilate phosphoribosyltransferase | | Authors: | Castell, A, Cookson, T.V.M, Bulloch, E, Evans, G.L, Baker, E.N, Lott, J.S, Parker, E.J. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Alternative substrates reveal catalytic cycle and key binding events in the reaction catalysed by anthranilate phosphoribosyltransferase from Mycobacterium tuberculosis.
Biochem.J., 461, 2014
|
|
4N31
 
 | |
4NWZ
 
 | | Structure of bacterial type II NADH dehydrogenase from Caldalkalibacillus thermarum at 2.5A resolution | | Descriptor: | FAD-dependent pyridine nucleotide-disulfide oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nakatani, Y, Heikal, A, Lott, J.S, Sazanov, L.A, Baker, E.N, Cook, G.M. | | Deposit date: | 2013-12-07 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the bacterial type II NADH dehydrogenase: a monotopic membrane protein with an essential role in energy generation.
Mol.Microbiol., 91, 2014
|
|
4NI6
 
 | |
