5DYJ
 
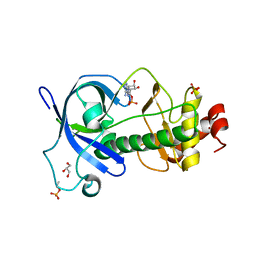 | | Mysosin heavy chain kinase A catalytic domain mutant - D663A | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Myosin heavy chain kinase A, ... | | Authors: | van Staalduinen, L.M, Yang, Y, Jia, Z. | | Deposit date: | 2015-09-24 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Dictyostelium Myosin-II Heavy Chain Kinase A (MHCK-A) alpha-kinase domain apoenzyme reveals a novel autoinhibited conformation.
Sci Rep, 6, 2016
|
|
5FDN
 
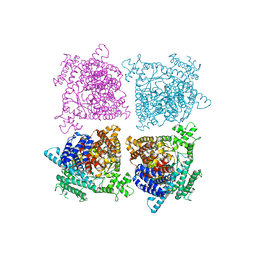 | |
3KCP
 
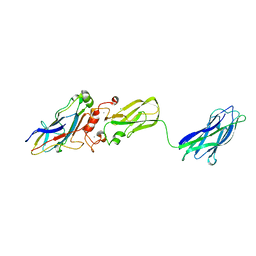 | | Crystal structure of interacting Clostridium thermocellum multimodular components | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cellulosomal-scaffolding protein A, ... | | Authors: | Adams, J.J, Currie, M.A, Bayer, E.A, Jia, Z, Smith, S.P. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into Higher-Order Organization of the Cellulosome Revealed by a Dissect-and-Build Approach: Crystal Structure of Interacting Clostridium thermocellum Multimodular Components
J.Mol.Biol., 396, 2010
|
|
3KMH
 
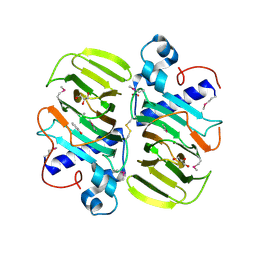 | |
3LCB
 
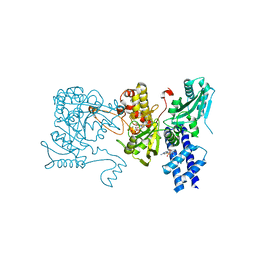 | | The crystal structure of isocitrate dehydrogenase kinase/phosphatase in complex with its substrate, isocitrate dehydrogenase, from Escherichia coli. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isocitrate dehydrogenase [NADP], ... | | Authors: | Zheng, J, Jia, Z. | | Deposit date: | 2010-01-10 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bifunctional isocitrate dehydrogenase kinase/phosphatase.
Nature, 465, 2010
|
|
3LMI
 
 | |
3LKM
 
 | |
3LMH
 
 | |
3LC6
 
 | |
3MPB
 
 | |
3LLA
 
 | |
6AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 M21A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
8MSI
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14SQ44T | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
9AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 S42G | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
4MSI
 
 | |
4IL1
 
 | | Crystal Structure of the Rat Calcineurin | | Descriptor: | CALCIUM ION, Calmodulin, Calcineurin subunit B type 1, ... | | Authors: | Ye, Q, Faucher, F, Jia, Z. | | Deposit date: | 2012-12-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of calcineurin activation by calmodulin.
Cell Signal, 25, 2013
|
|
4GAC
 
 | |
4FL4
 
 | | Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Glycoside hydrolase family 9, ... | | Authors: | Currie, M.A, Adams, J.J, Faucher, F, Bayer, E.A, Jia, Z, Smith, S.P, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
J.Biol.Chem., 287, 2012
|
|
4MF9
 
 | |
3TGM
 
 | | X-Ray Crystal Structure of Human Heme Oxygenase-1 in Complex with 1-(1H-imidazol-1-yl)-4,4-diphenyl-2 butanone | | Descriptor: | 1-(1H-imidazol-1-yl)-4,4-diphenylbutan-2-one, HEXANE-1,6-DIOL, Heme oxygenase 1, ... | | Authors: | Rahman, M.N, Jia, Z. | | Deposit date: | 2011-08-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A novel, "double-clamp" binding mode for human heme oxygenase-1 inhibition.
Plos One, 7, 2012
|
|
4ZME
 
 | |
4ZLH
 
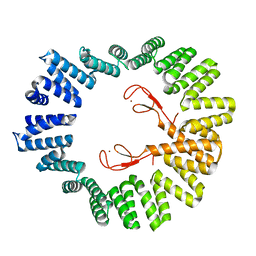 | | Structure of the LapB cytoplasmic domain at 2 angstroms | | Descriptor: | Lipopolysaccharide assembly protein B, ZINC ION | | Authors: | Prince, C.C, Jia, Z. | | Deposit date: | 2015-05-01 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Unexpected Duo: Rubredoxin Binds Nine TPR Motifs to Form LapB, an Essential Regulator of Lipopolysaccharide Synthesis.
Structure, 23, 2015
|
|
4ZMF
 
 | |
4ZS4
 
 | |
4NUB
 
 | |
