8BI8
 
 | | Structure of a cyclic beta-hairpin peptide derived from neuronal nitric oxide synthase | | Descriptor: | Nitric oxide synthase, brain | | Authors: | Balboa, J.R, Ostergaard, S, Stromgaard, K, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of a Potent Cyclic Peptide Inhibitor of the nNOS/PSD-95 Interaction.
J.Med.Chem., 66, 2023
|
|
8BI9
 
 | | Structure of a cyclic beta-hairpin peptide derived from neuronal nitric oxide synthase (T112W/T116E variant) | | Descriptor: | Nitric oxide synthase, brain, TERTIARY-BUTYL ALCOHOL | | Authors: | Balboa, J.R, Ostergaard, S, Stromgaard, K, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Development of a Potent Cyclic Peptide Inhibitor of the nNOS/PSD-95 Interaction.
J.Med.Chem., 66, 2023
|
|
4DIU
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR94 | | Descriptor: | Engineered Protein PF00326 | | Authors: | Seetharaman, J, Lew, S, Wang, D, Kohan, E, Patel, D, Whitehead, T, Fleishman, S, Ciccosanti, C, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR94
To be Published
|
|
4RHP
 
 | | Crystal structure of human COQ9 in complex with a phospholipid, Northeast Structural Genomics Consortium Target HR5043 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Ubiquinone biosynthesis protein COQ9, mitochondrial | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, H, Lee, D, Kogan, S, Maglaqui, M, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-22 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7Q8Z
 
 | | Crystal structure of TTBK2 in complex with VNG1.33 (compound 27) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 2, ~{N}-(4-phenoxyphenyl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
7Q90
 
 | | Crystal structure of TTBK2 in complex with VNG1.63 (compound 32) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 2, ~{N}-[4-(4-methoxyphenoxy)phenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
5ENF
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with fragment-4 N10142 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-2-(2-methylpropyl)-1,3-oxazole-4-carbonitrile, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Amin, J, Szykowska, A, Burgess-Brown, N, Spencer, J, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENH
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-12 N11528 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | PH-interacting protein, ~{N}-[(2,6-dimethoxyphenyl)methyl]ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENB
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with o-Tolylthiourea (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-methylphenyl)thiourea, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENI
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-13 N11537 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]-2-oxidanyl-ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
8A31
 
 | | p53 cancer mutant Y220C in complex with iodophenol-based small-molecule stabilizer JC694 | | Descriptor: | 4-(3-fluoranylpyrrol-1-yl)-3,5-bis(iodanyl)-2-oxidanyl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Balourdas, D.I, Stephenson Clarke, J.R, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C.
Acs Pharmacol Transl Sci, 5, 2022
|
|
8A32
 
 | | p53 cancer mutant Y220C in complex with iodophenol-based small-molecule stabilizer JC769 | | Descriptor: | 1,2-ETHANEDIOL, 4-[3,4-bis(fluoranyl)pyrrol-1-yl]-3,5-bis(iodanyl)-2-oxidanyl-benzoic acid, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Stephenson Clarke, J.R, Baud, M.G.J, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C.
Acs Pharmacol Transl Sci, 5, 2022
|
|
5ENC
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with N-(2,6-Dichlorobenzyl)acetamide (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
1NFQ
 
 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Androsterone, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-15 | | Release date: | 2002-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8BZJ
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MRLW5 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8BZI
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MR39 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2,5-bis(fluoranyl)-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
8C7Y
 
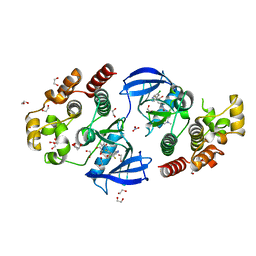 | | Crystal structure of BRAF V600E in complex with a hybrid compound 6 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, Serine/threonine-protein kinase B-raf, ... | | Authors: | Chaikuad, A, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis and characterisation of a novel type II B-RAF paradox breaker inhibitor.
Eur.J.Med.Chem., 250, 2023
|
|
4Z0R
 
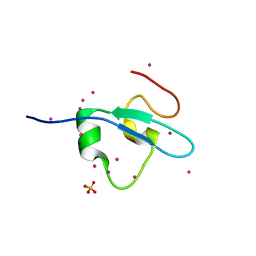 | | Crystal Structure of the CW domain of ZCWPW2 mutant F78R in complex with histone H3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Histone H3.1, SULFATE ION, ... | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the CW domain of ZCWPW2 mutant F78R in complex with histone H3 peptide
To be Published
|
|
1QXF
 
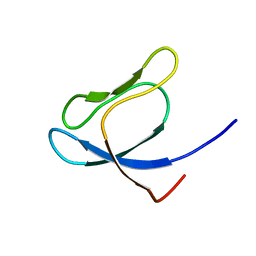 | | SOLUTION STRUCTURE OF 30S RIBOSOMAL PROTEIN S27E FROM ARCHAEOGLOBUS FULGIDUS: GR2, A NESG TARGET PROTEIN | | Descriptor: | 30S RIBOSOMAL PROTEIN S27E | | Authors: | Herve Du Penhoat, C, Atreya, H.S, Shen, Y, Liu, G, Acton, T.B, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the 30S ribosomal protein S27e encoded in gene RS27_ARCFU of Archaeoglobus fulgidis reveals a novel protein fold
Protein Sci., 13, 2004
|
|
1LXN
 
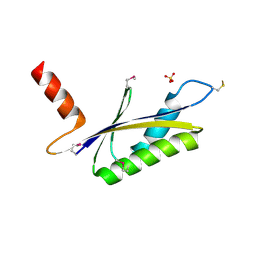 | | X-RAY STRUCTURE OF MTH1187 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT272 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1187, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of MTH1187 and its Yeast Ortholog YBL001C
Proteins, 52, 2003
|
|
4FFD
 
 | | Crystal structure of engineered protein. northeast structural genomics consortium target or48 | | Descriptor: | MAGNESIUM ION, PF00702 DOMAIN PROTEIN | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of engineered protein. northeast structural genomics consortium target or48
To be Published
|
|
4LOA
 
 | | X-ray structure of the de-novo design amidase at the resolution 1.8A, Northeast Structural Genomics Consortium (NESG) Target OR398 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, De-novo design amidase | | Authors: | Kuzin, A, Lew, S, Vorobiev, S.M, Seetharaman, J, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Khersonsky, O, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR398
To be Published
|
|
4PWW
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR494. | | Descriptor: | ACETIC ACID, OR494, PHOSPHATE ION | | Authors: | Vorobiev, S, Lin, Y.-R, Seetharaman, J, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Crystal Structure of Engineered Protein OR494.
To be Published
|
|
4XVX
 
 | | Crystal structure of an acyl-ACP dehydrogenase | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Acyl-[acyl-carrier-protein] dehydrogenase MbtN, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Chai, A, Johnston, J.M, Bunker, R.D, Lott, J.S, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-28 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A covalent adduct of MbtN, an acyl-ACP dehydrogenase from Mycobacterium tuberculosis, reveals an unusual acyl-binding pocket.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5C8G
 
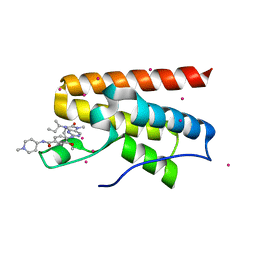 | | Crystal Structure Analysis of PP-BRD20 from Tb427tmp complexed with BI-2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, PP-BRD20, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, Hou, C.F.D, El Bakkouri, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Amain, M, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-25 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure Analysis of PP-BRD20 from Tb427tmp
to be published
|
|
