6SAQ
 
 | | wild-type NuoEF from Aquifex aeolicus bound to NADH-OH | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Gerhardt, S. | | Deposit date: | 2019-07-17 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis for Inhibition of ROS-Producing Respiratory Complex I by NADH-OH.
Angew.Chem.Int.Ed.Engl., 2021
|
|
8Q7C
 
 | | Cryo-EM structure of Adenovirus C5 hexon | | Descriptor: | Hexon protein | | Authors: | Zoll, S, Dhillon, A. | | Deposit date: | 2023-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the interaction between adenovirus C5 hexon and human lactoferrin.
J.Virol., 98, 2024
|
|
1WOF
 
 | | Crystal Structure Of SARS-CoV Mpro in Complex with an Inhibitor N1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Yang, H, Bartlam, M, Xue, X, Yang, K, Liang, W, Rao, Z. | | Deposit date: | 2004-08-18 | | Release date: | 2005-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
1B33
 
 | | STRUCTURE OF LIGHT HARVESTING COMPLEX OF ALLOPHYCOCYANIN ALPHA AND BETA CHAINS/CORE-LINKER COMPLEX AP*LC7.8 | | Descriptor: | ALLOPHYCOCYANIN, ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Reuter, W, Wiegand, G, Huber, R, Than, M.E. | | Deposit date: | 1998-12-15 | | Release date: | 1999-02-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis at 2.2 A of orthorhombic crystals presents the asymmetry of the allophycocyanin-linker complex, AP.LC7.8, from phycobilisomes of Mastigocladus laminosus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7LBM
 
 | | Structure of the human Mediator-bound transcription pre-initiation complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abdella, R, Talyzina, A, He, Y. | | Deposit date: | 2021-01-08 | | Release date: | 2021-03-24 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the human Mediator-bound transcription preinitiation complex.
Science, 372, 2021
|
|
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
7X2D
 
 | | Cryo-EM structure of the tavapadon-bound D1 dopamine receptor and mini-Gs complex | | Descriptor: | 1,5-dimethyl-6-[2-methyl-4-[3-(trifluoromethyl)pyridin-2-yl]oxy-phenyl]pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Teng, X, Zheng, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-15 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
7X2F
 
 | | Cryo-EM structure of the dopamine and LY3154207-bound D1 dopamine receptor and mini-Gs complex | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Teng, X, Zheng, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
7X2C
 
 | | Cryo-EM structure of the fenoldopam-bound D1 dopamine receptor and mini-Gs complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Teng, X, Zheng, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
6NHW
 
 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NHY
 
 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
8AB5
 
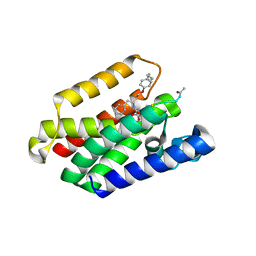 | | Structure of E. coli GlpG in complex with peptide derived inhibitor Ac-VRHA-conh-[4-(4-butyl)-phenoxy-1-phenyl-2-butyl] | | Descriptor: | Ac-VRHA-conh-[4-(4-butyl)-phenoxy-1-phenyl-2-butyl], Rhomboid protease GlpG | | Authors: | Skerlova, J, Polovinkin, V, Bach, K, Borshchevskiy, V, Strisovsky, K. | | Deposit date: | 2022-07-04 | | Release date: | 2023-10-25 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Extensive targeting of chemical space at the prime side of ketoamide inhibitors of rhomboid proteases by branched substituents empowers their selectivity and potency.
Eur.J.Med.Chem., 275, 2024
|
|
6RJK
 
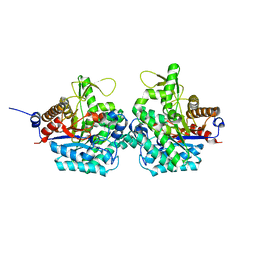 | | Structure of virulence factor SghA from Agrobacterium tumefaciens | | Descriptor: | Beta-glucosidase | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8DVH
 
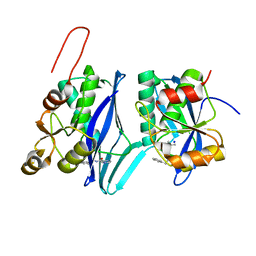 | | Crystal structure of ATP-dependent Lon protease from Bacillus subtillis (BsLonBA) | | Descriptor: | Lon protease 2, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, SODIUM ION | | Authors: | Sekula, B, Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unique Structural Fold of LonBA Protease from Bacillus subtilis, a Member of a Newly Identified Subfamily of Lon Proteases.
Int J Mol Sci, 23, 2022
|
|
8B8S
 
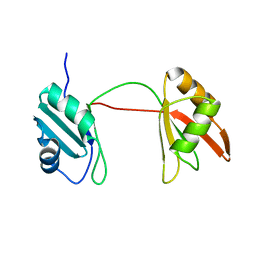 | | Solution structure of tandem RRM1 and RRM2 domains of yeast NPL3 | | Descriptor: | Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 | | Authors: | Kachariya, N, Sattler, M, Keil, P, Strasser, K. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res., 51, 2023
|
|
7XMK
 
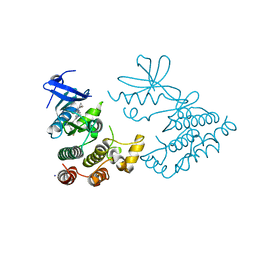 | | Crystal structure of human RIPK1 kinase domain in complex with compound SKLB923 | | Descriptor: | 5-[2-(cyclopropylcarbonylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-(3-fluorophenyl)ethyl]-1-methyl-indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Yang, S. | | Deposit date: | 2022-04-26 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | From Hit to Lead: Structure-Based Optimization of Novel Selective Inhibitors of Receptor-Interacting Protein Kinase 1 (RIPK1) for the Treatment of Inflammatory Diseases.
J.Med.Chem., 67, 2024
|
|
7MWH
 
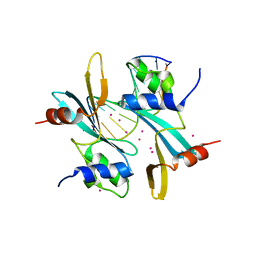 | | Crystal structure of BAZ2A with DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the BAZ2B TAM domain.
Heliyon, 8, 2022
|
|
5DW3
 
 | |
7LDD
 
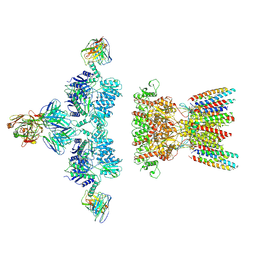 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
5DVZ
 
 | | Holo TrpB from Pyrococcus furiosus | | Descriptor: | PHOSPHATE ION, SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Buller, A.R, Arnold, F.H. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-03 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Directed evolution of the tryptophan synthase beta-subunit for stand-alone function recapitulates allosteric activation.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6GOU
 
 | | Development of Alkyl Glycerone Phosphate Synthase Inhibitors: Complex with Inhibitor 2I | | Descriptor: | (3~{S})-3-[2,6-bis(fluoranyl)phenyl]-~{N}-[(2-oxidanylidene-1,3-dihydrobenzimidazol-5-yl)methyl]butanamide, Alkyldihydroxyacetonephosphate synthase, peroxisomal, ... | | Authors: | Mattevi, A, Piano, V. | | Deposit date: | 2018-06-04 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of alkyl glycerone phosphate synthase inhibitors: Structure-activity relationship and effects on ether lipids and epithelial-mesenchymal transition in cancer cells.
Eur J Med Chem, 163, 2018
|
|
6RJM
 
 | | Complex structure of virulence factor SghA and its hydrolysis product glucose | | Descriptor: | Beta-glucosidase, alpha-D-glucopyranose | | Authors: | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2019-04-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6I2U
 
 | |
