9IQF
 
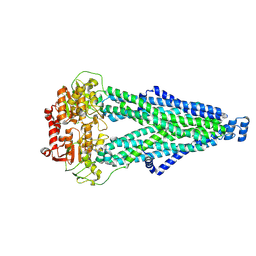 | |
9IQG
 
 | | Cryo-EM structure of MsRv1273c/72c from Mycobacterium smegmatis in the ATP|ADP+Vi-bound Occ (Vi) state | | Descriptor: | ABC transporter transmembrane region, ABC transporter, ATP-binding protein, ... | | Authors: | Lan, Y, Yu, J, Li, J. | | Deposit date: | 2024-07-12 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of a mycobacterial isoniazid efflux pump MsRv1273c/72c with a degenerate nucleotide-binding site.
Nat Commun, 16, 2025
|
|
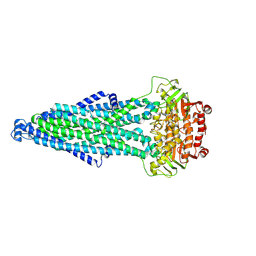
8H41
 
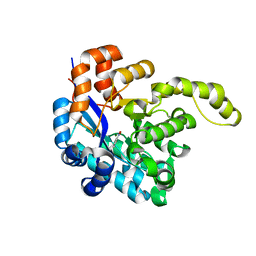 | | Crystal structure of a decarboxylase from Trichosporon moniliiforme in complex with o-nitrophenol | | Descriptor: | MAGNESIUM ION, O-NITROPHENOL, Salicylate decarboxylase | | Authors: | Gao, J, Zhao, Y.P, Li, Q, Liu, W.D, Sheng, X. | | Deposit date: | 2022-10-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Combined Computational-Experimental Study on the Substrate Binding and Reaction Mechanism of Salicylic Acid Decarboxylase
Catalysts, 12, 2022
|
|
7YOJ
 
 | | Structure of CasPi with guide RNA and target DNA | | Descriptor: | CasPi, DNA (30-MER), DNA (5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*CP*CP*AP*G)-3'), ... | | Authors: | Li, C.P, Wang, J, Liu, J.J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The compact Cas pi (Cas12l) 'bracelet' provides a unique structural platform for DNA manipulation.
Cell Res., 33, 2023
|
|
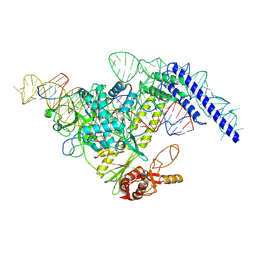
7YRV
 
 | |
7YRW
 
 | |
7YRX
 
 | |
6Z2M
 
 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 antibody, Spike glycoprotein, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-17 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published
|
|
6YZ7
 
 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Cr3022, Antibody light chain, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published, 2020
|
|
9IM1
 
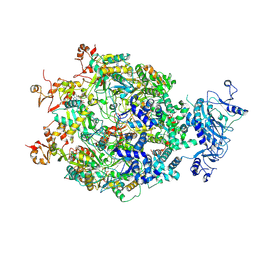 | | The Cryo-EM structure of MPXV E5 in complex with ssDNA in intermediate state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Cheng, Y.X, Han, P, Wang, H. | | Deposit date: | 2024-07-01 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Assembly and breakage of head-to-head double hexamer reveals mpox virus E5-catalyzed DNA unwinding initiation.
Nat Commun, 16, 2025
|
|
9IM0
 
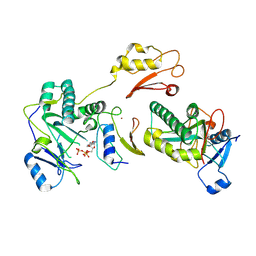 | |
9ILY
 
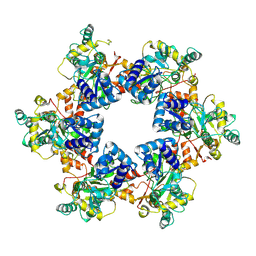 | |
9ILZ
 
 | | The Cryo-EM structure of MPXV E5 in complex with ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Cheng, Y.X, Han, P, Wang, H. | | Deposit date: | 2024-07-01 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Assembly and breakage of head-to-head double hexamer reveals mpox virus E5-catalyzed DNA unwinding initiation.
Nat Commun, 16, 2025
|
|
9IM2
 
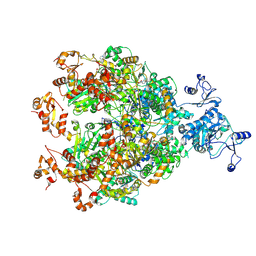 | | The Cryo-EM structure of MPXV E5 in complex with ssDNA in intermediate state 3 | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Cheng, Y.X, Han, P, Wang, H. | | Deposit date: | 2024-07-01 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Assembly and breakage of head-to-head double hexamer reveals mpox virus E5-catalyzed DNA unwinding initiation.
Nat Commun, 16, 2025
|
|
7CCD
 
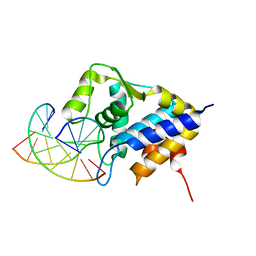 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*GP*AS*AP*CP*GP*TP*G)-3'), HNHc domain-containing protein | | Authors: | Yu, H, Li, J, Liu, G, Zhao, G, Wang, Y, Hu, W, Deng, Z, Gan, J, Zhao, Y, He, X. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
8XY6
 
 | | ASFV RNAP M1249L C-tail occupied complex3 (MCOC3) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XXT
 
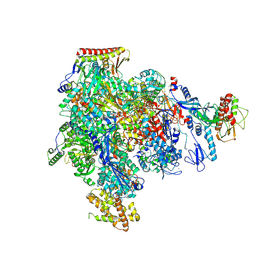 | | ASFV RNAP M1249L C-tail occupied complex2 (MCOC2) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XX4
 
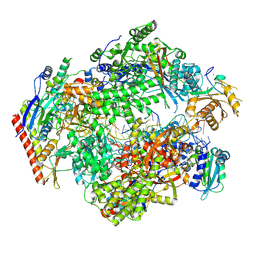 | | ASFV RNAP elongation complex | | Descriptor: | C122R, D339L, DNA (5'-D(P*CP*GP*CP*AP*AP*CP*GP*GP*CP*GP*A)-3'), ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XX5
 
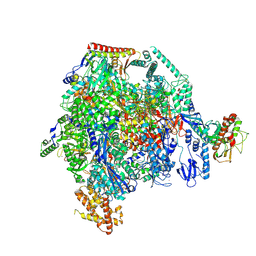 | | ASFV RNAP M1249L C-tail occupied complex1 (MCOC1) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XXP
 
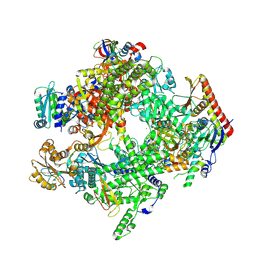 | | ASFV RNAP core complex | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8Y0E
 
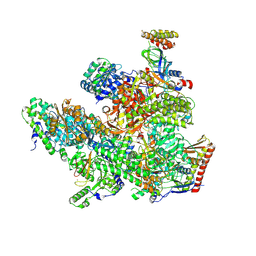 | | ASFV RNAP M1249L C-tail occupied complex4 (MCOC4) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-22 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
9IK4
 
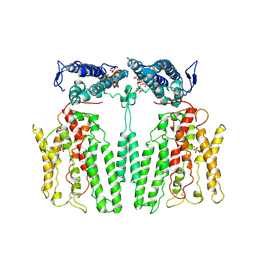 | |
9JF8
 
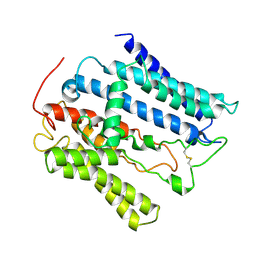 | |
4KMA
 
 | |
9J8V
 
 | | TSWV L protein in complex with ribavirin 5-triphosphate | | Descriptor: | RIBAVIRIN TRIPHOSPHATE, RNA-directed RNA polymerase L | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2024-08-21 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
