6XH2
 
 | |
4XNN
 
 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Daphnia pulex | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL | | Authors: | Ebrahim, A, Hobdey, S.E, Podkaminer, K, Taylor II, L.E, Beckham, G.T, Decker, S.R, Himmel, M.E, Cragg, S.M, McGeehan, J.E. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a GH7 Family Cellobiohydrolase from Daphnia pulex
To Be Published
|
|
2NUF
 
 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 2.5-Angstrom Resolution | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
3LOB
 
 | | Crystal Structure of Flock House Virus calcium mutant | | Descriptor: | Coat protein beta, Coat protein gamma, RNA (5'-R(*UP*UP*U*AP*UP*CP*UP*(P))-3'), ... | | Authors: | Johnson, J.E, Banerjee, M, Speir, J.A, Huang, R. | | Deposit date: | 2010-02-03 | | Release date: | 2010-04-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and function of a genetically engineered mimic of a nonenveloped virus entry intermediate.
J.Virol., 84, 2010
|
|
3LCP
 
 | | Crystal structure of the carbohydrate recognition domain of LMAN1 in complex with MCFD2 | | Descriptor: | CALCIUM ION, Multiple coagulation factor deficiency protein 2, Protein ERGIC-53 | | Authors: | Wigren, E, Bourhis, J.M, Kursula, I, Guy, J.E, Lindqvist, Y. | | Deposit date: | 2010-01-11 | | Release date: | 2010-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the LMAN1-CRD/MCFD2 transport receptor complex provides insight into combined deficiency of factor V and factor VIII.
Febs Lett., 584, 2010
|
|
2QJJ
 
 | | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
4Y2E
 
 | | Crystal structure of the catalytic domain of human dual-specificity phosphatase 7 (C232S) | | Descriptor: | Dual specificity protein phosphatase 7, PHOSPHATE ION | | Authors: | Lountos, G.T, Austin, B.P, Tropea, J.E, Waugh, D.S. | | Deposit date: | 2015-02-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of human dual-specificity phosphatase 7, a potential cancer drug target.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2QBX
 
 | | EphB2/SNEW Antagonistic Peptide Complex | | Descriptor: | Ephrin type-B receptor 2, SULFATE ION, antagonistic peptide | | Authors: | Chrencik, J.E, Brooun, A, Recht, M.I, Nicola, G, Pasquale, E.B, Kuhn, P, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-06-18 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the EphB2 receptor in complex with an antagonistic peptide reveals a novel mode of inhibition.
J.Biol.Chem., 282, 2007
|
|
4XYP
 
 | | Crystal structure of a piscine viral fusion protein | | Descriptor: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, CHLORIDE ION, Fusion protein | | Authors: | Cook, J.D, Lee, J.E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Electrostatic Architecture of the Infectious Salmon Anemia Virus (ISAV) Core Fusion Protein Illustrates a Carboxyl-Carboxylate pH Sensor.
J.Biol.Chem., 290, 2015
|
|
2OT5
 
 | |
2OUE
 
 | |
6XMJ
 
 | | Human 20S proteasome bound to an engineered 11S (PA26) activator | | Descriptor: | Proteasome activator protein PA26, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | de la Pena, A.H, Opoku-Nsiah, K.A, Williams, S.K, Chopra, N, Sali, A, Gestwicki, J.E, Lander, G.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Y Phi motif defines the structure-activity relationships of human 20S proteasome activators.
Nat Commun, 13, 2022
|
|
6XH1
 
 | |
1YPT
 
 | | CRYSTAL STRUCTURE OF YERSINIA PROTEIN TYROSINE PHOSPHATASE AT 2.5 ANGSTROMS AND THE COMPLEX WITH TUNGSTATE | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YERSINIA (CATALYTIC DOMAIN) | | Authors: | Stuckey, J.A, Schubert, H.L, Fauman, E.B, Zhang, Z.-Y, Dixon, J.E, Saper, M.A. | | Deposit date: | 1994-09-16 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate.
Nature, 370, 1994
|
|
4YG8
 
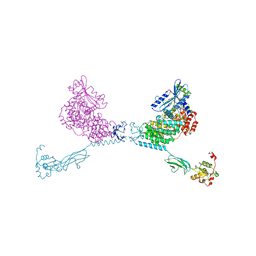 | | CRYSTAL STRUCTURE OF THE CHS5-CHS6 EXOMER CARGO ADAPTOR COMPLEX | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Chitin biosynthesis protein CHS5, Chitin biosynthesis protein CHS6, ... | | Authors: | Paczkowski, J.E, Richardson, B.C, Strassner, A.M, Fromme, J.C. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The exomer cargo adaptor structure reveals a novel GTPase-binding domain.
Embo J., 31, 2012
|
|
4YCA
 
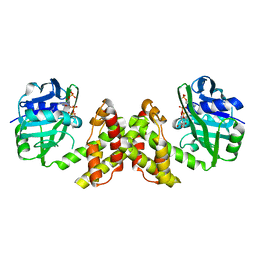 | | Evidence of Kinetic Cooperativity in dimeric Ketopantoate Reductase from Staphylococcus aureus | | Descriptor: | 2-dehydropantoate 2-reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sanchez, J.E, Gross, P.G, Goetze, R, Walsh Jr, R.M, Peeples, W.B, Wood, Z.A. | | Deposit date: | 2015-02-19 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Evidence of Kinetic Cooperativity in Dimeric Ketopantoate Reductase from Staphylococcus aureus.
Biochemistry, 54, 2015
|
|
3LX2
 
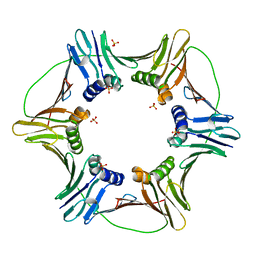 | |
6Y5S
 
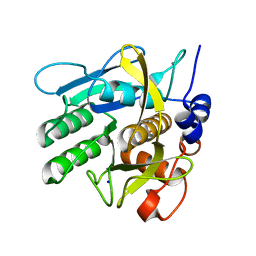 | | Crystal structure of savinase at cryogenic conditions | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
1YVL
 
 | | Structure of Unphosphorylated STAT1 | | Descriptor: | 5-residue peptide, GOLD ION, Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Mao, X, Ren, Z, Parker, G.N, Sondermann, H, Pastorello, M.A, Wang, W, McMurray, J.S, Demeler, B, Darnell Jr, J.E, Chen, X. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural bases of unphosphorylated STAT1 association and receptor binding.
Mol.Cell, 17, 2005
|
|
6Y6T
 
 | | Mouse Galactocerebrosidase complexed with galacto-noeurostegine GNS at pH 4.6 | | Descriptor: | (1~{R},2~{S},3~{S},4~{R},5~{R})-4-(hydroxymethyl)-8-azabicyclo[3.2.1]octane-1,2,3-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Deane, J.E, McLoughlin, J. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Bicyclic Form of galacto -Noeurostegine Is a Potent Inhibitor of beta-Galactocerebrosidase.
Acs Med.Chem.Lett., 12, 2021
|
|
2QJN
 
 | | Crystal structure of D-mannonate dehydratase from Novosphingobium aromaticivorans complexed with Mg and 2-keto-3-deoxy-D-gluconate | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
2BDO
 
 | | SOLUTION STRUCTURE OF HOLO-BIOTINYL DOMAIN FROM ACETYL COENZYME A CARBOXYLASE OF ESCHERICHIA COLI DETERMINED BY TRIPLE-RESONANCE NMR SPECTROSCOPY | | Descriptor: | BIOTIN, PROTEIN (ACETYL-COA CARBOXYLASE) | | Authors: | Roberts, E.L, Shu, N, Howard, M.J, Broadhurst, R.W, Chapman-Smith, A, Wallace, J.C, Morris, T, Cronan, J.E, Perham, R.N. | | Deposit date: | 1999-03-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
2BCZ
 
 | | Crystal Structure of a minimal, mutant all-RNA hairpin ribozyme (U39C, G8I, 2'deoxy A-1) | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*IP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(DA)P*GP*UP*CP*CP*AP*CP*CP*G)-3'), ... | | Authors: | Salter, J.D, Wedekind, J.E. | | Deposit date: | 2005-10-19 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Water in the Active Site of an All-RNA Hairpin Ribozyme and Effects of Gua8 Base Variants on the Geometry of Phosphoryl Transfer.
Biochemistry, 45, 2006
|
|
4WOH
 
 | | Structure of of human dual-specificity phosphatase 22 (E24A/K28A/K30A/C88S) complexed with 4-nitrophenolphosphate | | Descriptor: | 1,2-ETHANEDIOL, 4-NITROPHENYL PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Cherry, S, Tropea, J.E, Waugh, D.S. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural analysis of human dual-specificity phosphatase 22 complexed with a phosphotyrosine-like substrate.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2QBY
 
 | | Crystal structure of a heterodimer of Cdc6/Orc1 initiators bound to origin DNA (from S. solfataricus) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 6 homolog 1, Cell division control protein 6 homolog 3, ... | | Authors: | Cunningham Dueber, E.L, Corn, J.E, Bell, S.D, Berger, J.M. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex.
Science, 317, 2007
|
|
