8AQK
 
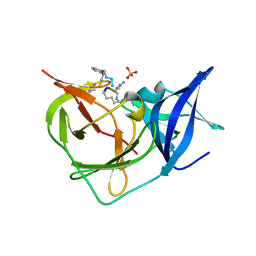 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2258 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, SULFATE ION, Serine protease NS3, ... | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.
Arch Pharm, 2024
|
|
8AQB
 
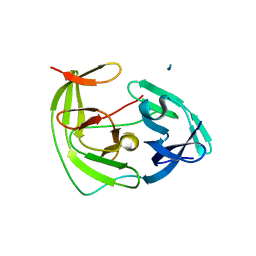 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2257 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, ACETATE ION, Serine protease NS3, ... | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-08-11 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.
Arch Pharm, 2024
|
|
8AQA
 
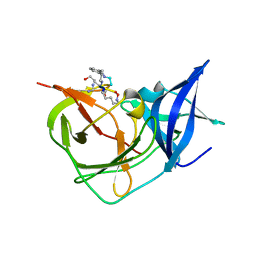 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2260 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-08-11 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.
Arch Pharm, 2024
|
|
4I77
 
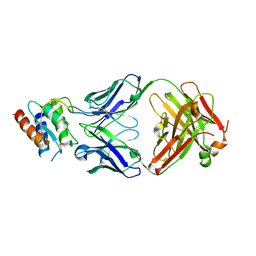 | | Lebrikizumab Fab bound to IL-13 | | Descriptor: | Interleukin-13, Lebrikizumab heavy chain, Lebrikizumab light chain | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Signaling Blockade by Anti-IL-13 Antibody Lebrikizumab.
J.Mol.Biol., 425, 2013
|
|
4JQX
 
 | | HLA-B*44:03 in complex with Epstein-Barr virus BZLF1-derived peptide (residues 169-180) | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Theodossis, A, Welland, A, Gras, S, Rossjohn, J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HLA Peptide Length Preferences Control CD8+ T Cell Responses.
J.Immunol., 191, 2013
|
|
4JQV
 
 | | HLA-B*18:01 in complex with Epstein-Barr virus BZLF1-derived peptide (residues 173-180) | | Descriptor: | ACETATE ION, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Welland, A, Gras, S, Rossjohn, J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | HLA Peptide Length Preferences Control CD8+ T Cell Responses.
J.Immunol., 191, 2013
|
|
4K25
 
 | | Crystal Structure of yeast Qri7 homodimer | | Descriptor: | CALCIUM ION, Probable tRNA threonylcarbamoyladenosine biosynthesis protein QRI7, mitochondrial, ... | | Authors: | Neculai, D, Wan, L, Mao, D.Y, Sicheri, F. | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-08 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system.
Nucleic Acids Res., 41, 2013
|
|
4LED
 
 | | The Crystal Structure of Pyocin L1 bound to D-rhamnose at 2.37 Angstroms | | Descriptor: | Pyocin L1, alpha-D-rhamnopyranose | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4LEA
 
 | | The Crystal Structure of Pyocin L1 bound to D-mannose at 2.55 Angstroms | | Descriptor: | Pyocin L1, beta-D-mannopyranose | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4L1A
 
 | | Crystallographic study of multi-drug resistant HIV-1 protease Lopinavir complex: mechanism of drug recognition and resistance | | Descriptor: | MDR769 HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE | | Authors: | Liu, Z, Yedidi, R.S, Wang, Y, Dewdney, T, Reiter, S, Brunzelle, J, Kovari, I, Kovari, L. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of multi-drug resistant HIV-1 protease lopinavir complex: mechanism of drug recognition and resistance.
Biochem.Biophys.Res.Commun., 437, 2013
|
|
4LE7
 
 | | The Crystal Structure of Pyocin L1 at 2.09 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Pyocin L1 | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, R.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
7LII
 
 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S7D | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIL
 
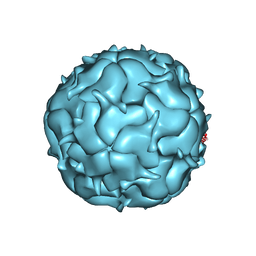 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant SE3 | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIM
 
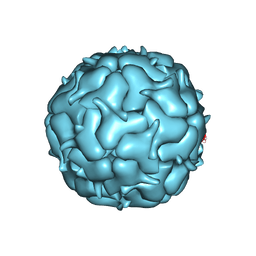 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S6E | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIJ
 
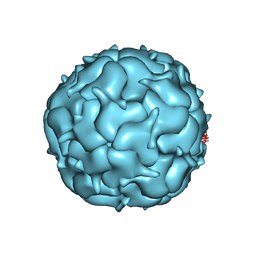 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S1K | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIK
 
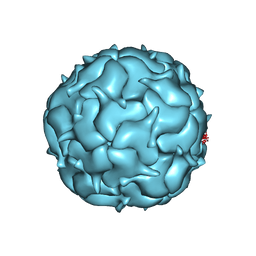 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S1R | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIS
 
 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S5D | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIT
 
 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S7G | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7MU9
 
 | |
7MHC
 
 | | Structure of human STING in complex with MK-1454 | | Descriptor: | (2R,5R,7R,8S,10R,12aR,14R,15S,15aR,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-15,16-difluoro-2,10-bis(sulfanyl)octahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A kinase-cGAS cascade to synthesize a therapeutic STING activator.
Nature, 603, 2022
|
|
7N5S
 
 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 6) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
7N5T
 
 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 5) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
7NPF
 
 | | Vibrio cholerae ParA2-ATPyS-DNA filament | | Descriptor: | AAA family ATPase, DNA (49-MER), MAGNESIUM ION, ... | | Authors: | Parker, A.V, Bergeron, J.R.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The structure of the bacterial DNA segregation ATPase filament reveals the conformational plasticity of ParA upon DNA binding.
Nat Commun, 12, 2021
|
|
7NLV
 
 | | WILDTYPE CORE-STREPTAVIDIN WITH a conjugated BIOTINYLATED PYRROLIDINE II | | Descriptor: | 5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-N-((S)-pyrrolidin-3-yl)pentanamide, Streptavidin | | Authors: | Nodling, A.R, Santi, N, Tsai, Y.H, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The role of streptavidin and its variants in catalysis by biotinylated secondary amines.
Org.Biomol.Chem., 19, 2021
|
|
7NPD
 
 | | Vibiro cholerae ParA2 | | Descriptor: | Walker A-type ATPase | | Authors: | Parker, A.V, Bergeron, J.R.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the bacterial DNA segregation ATPase filament reveals the conformational plasticity of ParA upon DNA binding.
Nat Commun, 12, 2021
|
|
