4E1J
 
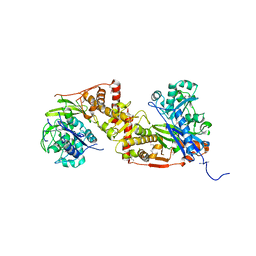 | | Crystal structure of glycerol kinase in complex with glycerol from Sinorhizobium meliloti 1021 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycerol kinase, ... | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-06 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of glycerol kinase in complex with glycerol from Sinorhizobium meliloti 1021
To be Published
|
|
3IJ6
 
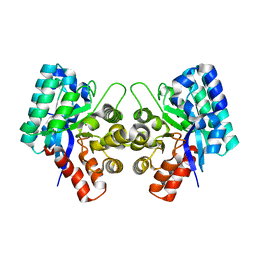 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidophilus | | Descriptor: | SODIUM ION, UNCHARACTERIZED METAL-DEPENDENT HYDROLASE, ZINC ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-03 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidopphilus
To be Published
|
|
3ID7
 
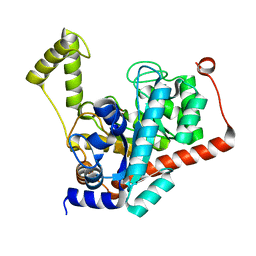 | | Crystal structure of renal dipeptidase from Streptomyces coelicolor A3(2) | | Descriptor: | CHLORIDE ION, Dipeptidase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-07-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
4DXK
 
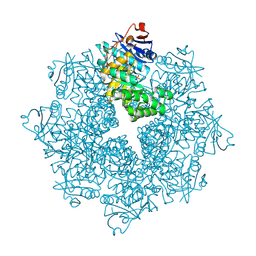 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue, na and phosphate | | Descriptor: | Mandelate racemase / muconate lactonizing enzyme family protein, PHOSPHATE ION, SODIUM ION | | Authors: | Vetting, M.W, Bouvier, J.T, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-27 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue, na and phosphate
to be published
|
|
4DZI
 
 | | Crystal structure of amidohydrolase map2389c (target EFI-500390) from Mycobacterium avium subsp. paratuberculosis K-10 | | Descriptor: | CADMIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of amidohydrolase map2389c (target EFI-500390) from Mycobacterium avium
To be Published
|
|
3IOY
 
 | |
4DYV
 
 | | Crystal structure of a short-chain dehydrogenase/reductase SDR from Xanthobacter autotrophicus Py2 | | Descriptor: | CHLORIDE ION, Short-chain dehydrogenase/reductase SDR | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Gizzi, A, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a short-chain dehydrogenase/reductase SDR from Xanthobacter autotrophicus Py2
To be Published
|
|
4DYK
 
 | | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn | | Descriptor: | AMIDOHYDROLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Raushel, F.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an adenosine deaminase from pseudomonas aeruginosa pao1 (target nysgrc-200449) with bound zn
to be published
|
|
4DLF
 
 | | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P3221 | | Descriptor: | Amidohydrolase 2, GLYCEROL, UNKNOWN ATOM OR ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (target efi-500235) with bound ZN, space group P3221
to be published
|
|
4DPO
 
 | | Crystal structure of a conserved protein MM_1583 from Methanosarcina mazei Go1 | | Descriptor: | Conserved protein | | Authors: | Agarwal, R, Chamala, S, Evans, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Foti, R, Siedel, R, Zencheck, W, Villigas, G, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-13 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of a conserved protein MM_1583 from Methanosarcina mazei Go1
To be Published
|
|
3GHY
 
 | | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2 | | Descriptor: | Ketopantoate reductase protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2
To be Published
|
|
4DWO
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Vetting, M.W, Wasserman, S.R, Morisco, L.L, Sojitra, S, Allen, K.N, Dunaway-Mariano, D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-26 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II
To be Published
|
|
4DQX
 
 | | Crystal structure of a short chain dehydrogenase from Rhizobium etli CFN 42 | | Descriptor: | Probable oxidoreductase protein | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a short chain dehydrogenase from Rhizobium etli CFN 42
To be Published
|
|
4DBD
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Sulfolobus solfataricus | | Descriptor: | Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-14 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Sulfolobus solfataricus
To be Published
|
|
4DF0
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Thermoproteus neutrophilus | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-22 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Thermoproteus neutrophilus
To be Published
|
|
3G1X
 
 | | Crystal structure of the mutant D70G of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | CHLORIDE ION, Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3GFG
 
 | | Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form | | Descriptor: | Uncharacterized oxidoreductase yvaA | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Chang, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form.
To be published
|
|
4DN2
 
 | | CRYSTAL STRUCTURE OF putative Nitroreductase from Geobacter metallireducens GS-15 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-08 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF putative Nitroreductase from Geobacter metallireducens GS-15
To be Published
|
|
3FVD
 
 | | Crystal structure of a member of enolase superfamily from ROSEOVARIUS NUBINHIBENS ISM complexed with magnesium | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Malashkevich, V.N, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a member of enolase superfamily from ROSEOVARIUS NUBINHIBENS ISM complexed with magnesium
to be published
|
|
4DNG
 
 | | Crystal structure of putative aldehyde dehydrogenase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Uncharacterized aldehyde dehydrogenase AldY | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative aldehyde dehydrogenase from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
3FYF
 
 | | Crystal structure of uncharacterized protein bvu_3222 from bacteroides vulgatus | | Descriptor: | PROTEIN BVU-3222 | | Authors: | Patskovsky, Y, Bonanno, J.B, Ozyurt, S, Rutter, M, Chang, S, Groshong, C, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Protein Bvu-3222 from Bacteroides Vulgatus
To be Published
|
|
3FYY
 
 | | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with Mg | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
3G12
 
 | | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus | | Descriptor: | Putative lactoylglutathione lyase, SULFATE ION | | Authors: | Patskovsky, Y, Madegowda, M, Gilmore, M, Chang, S, Maletic, M, Smith, D, Sauder, J.M, Burley, S.K, Swaminathan, S, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus
To be Published
|
|
3G1D
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
4DLL
 
 | | Crystal structure of a 2-hydroxy-3-oxopropionate reductase from Polaromonas sp. JS666 | | Descriptor: | 2-hydroxy-3-oxopropionate reductase, SULFATE ION | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of a 2-hydroxy-3-oxopropionate reductase from Polaromonas sp. JS666
To be Published
|
|
