2ZZN
 
 | | The complex structure of aTrm5 and tRNACys | | Descriptor: | MAGNESIUM ION, RNA (71-MER), S-ADENOSYLMETHIONINE, ... | | Authors: | Goto-Ito, S, Ito, T, Yokoyama, S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3A5Q
 
 | | Benzalacetone synthase from Rheum palmatum | | Descriptor: | Benzalacetone synthase | | Authors: | Morita, H, Kato, R, Abe, I, Sugio, S, Kohno, T. | | Deposit date: | 2009-08-10 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for benzalacetone synthase from Rheum palmatum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A5S
 
 | | Benzalacetone synthase (I207L/L208F) | | Descriptor: | Benzalacetone synthase | | Authors: | Morita, H, Kato, R, Abe, I, Sugio, S, Kohno, T. | | Deposit date: | 2009-08-10 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for benzalacetone synthase from Rheum palmatum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2ZZM
 
 | | The complex structure of aTrm5 and tRNALeu | | Descriptor: | MAGNESIUM ION, RNA (84-MER), S-ADENOSYLMETHIONINE, ... | | Authors: | Goto-Ito, S, Ito, T, Yokoyama, S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation
Nat.Struct.Mol.Biol., 16, 2009
|
|
2ZPX
 
 | |
3WQ5
 
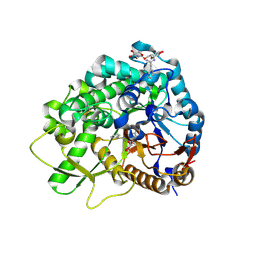 | |
2ZJC
 
 | | TNFR1 selectve TNF mutant; R1-6 | | Descriptor: | GLYCEROL, Tumor necrosis factor | | Authors: | Mukai, Y, Yamagata, Y, Tsutsumi, Y. | | Deposit date: | 2008-03-05 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Function Relationship of Tumor Necrosis Factor (TNF) and Its Receptor Interaction Based on 3D Structural Analysis of a Fully Active TNFR1-Selective TNF Mutant
J.Mol.Biol., 385, 2009
|
|
3WQ6
 
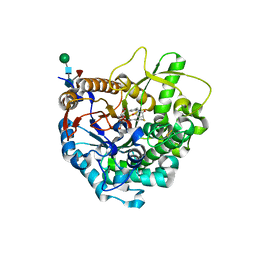 | |
3WQ4
 
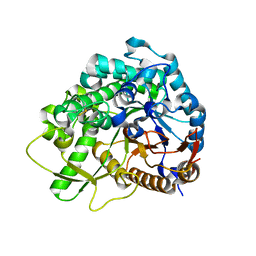 | | Crystal structure of beta-primeverosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-primeverosidase | | Authors: | Saino, H. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of beta-primeverosidase in complex with disaccharide amidine inhibitors.
J.Biol.Chem., 289, 2014
|
|
7COH
 
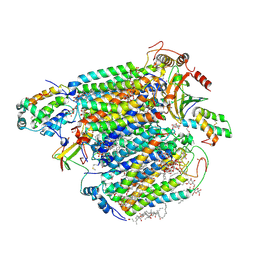 | | Dimeric Form of Bovine Heart Cytochrome c Oxidase in the Fully Oxidized State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Shinzawa-Itoh, K, Muramoto, K. | | Deposit date: | 2020-08-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3-A Resolution structure of bovine cytochrome c oxidase suggests a dimerization mechanism
Biochim.Biophys.Acta, 2021
|
|
4MT9
 
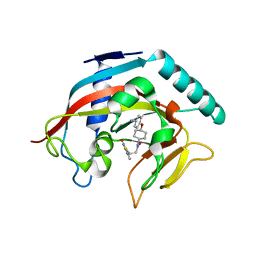 | | Co-crystal structure of tankyrase 1 with compound 49 | | Descriptor: | N-[trans-4-(4-cyanophenoxy)cyclohexyl]-3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-19 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4MSK
 
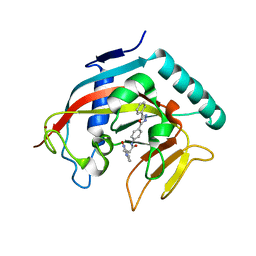 | | Co-crystal structure of tankyrase 1 with compound 34 | | Descriptor: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[4-(5-phenyl-1,3,4-oxadiazol-2-yl)phenyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4OBP
 
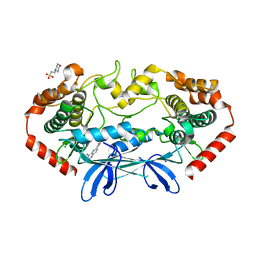 | | MAP4K4 in complex with inhibitor (compound 29), 6-(2-FLUOROPYRIDIN-4-YL)PYRIDO[3,2-D]PYRIMIDIN-4-AMINE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-fluoropyridin-4-yl)pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2014-01-07 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of Selective 4-Amino-pyridopyrimidine Inhibitors of MAP4K4 Using Fragment-Based Lead Identification and Optimization.
J.Med.Chem., 57, 2014
|
|
4MSG
 
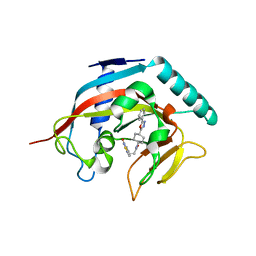 | | Crystal structure of tankyrase 1 with compound 22 | | Descriptor: | 3-[(4-oxo-3,4-dihydroquinazolin-2-yl)sulfanyl]-N-[trans-4-(5-phenyl-1,3,4-oxadiazol-2-yl)cyclohexyl]propanamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel dual binders as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
7ERO
 
 | | Crystal structure of D-allulose 3-epimerase with D-allulose from Agrobacterium sp. SUL3 | | Descriptor: | D-psicose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
7ERN
 
 | | Crystal structure of D-allulose 3-epimerase with D-fructose from Agrobacterium sp. SUL3 | | Descriptor: | D-fructose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
7ERM
 
 | | Crystal structure of D-allulose 3-epimerase from Agrobacterium sp. SUL3 | | Descriptor: | D-tagatose 3-epimerase, MAGNESIUM ION, SULFATE ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
