3KEB
 
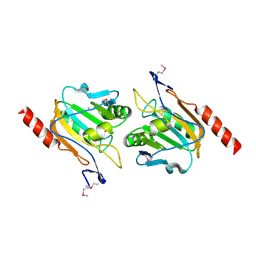 | | Thiol peroxidase from Chromobacterium violaceum | | Descriptor: | CHLORIDE ION, Probable thiol peroxidase, SULFATE ION | | Authors: | Osipiuk, J, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-25 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of thiol peroxidase from Chromobacterium violaceum
To be Published
|
|
3KKD
 
 | | Structure of a putative tetr transcriptional regulator (pa3699) from pseudomonas aeruginosa pa01 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Filippova, E.V, Chruszcz, M, Cymborowski, M, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-05 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Putative TetR Transcriptional Regulator (PA3699) from Pseudomonas Aeruginosa PA01
To be Published
|
|
3HBC
 
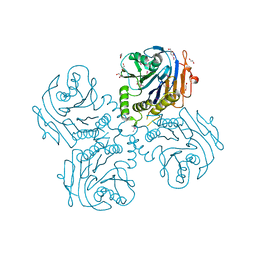 | | Crystal Structure of Choloylglycine Hydrolase from Bacteroides thetaiotaomicron VPI | | Descriptor: | 1,2-ETHANEDIOL, Choloylglycine hydrolase, GLYCEROL | | Authors: | Kim, Y, Bigelow, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Crystal Structure of Choloylglycine Hydrolase from Bacteroides thetaiotaomicron VPI
To be Published
|
|
3GNJ
 
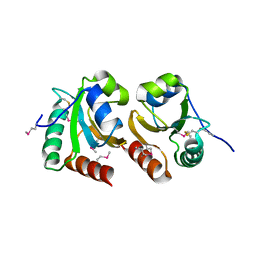 | | The crystal structure of a thioredoxin-related protein from Desulfitobacterium hafniense DCB | | Descriptor: | Thioredoxin domain protein | | Authors: | Tan, K, Volkart, L, Gu, M, Kinney, J.N, Babnigg, G, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of a thioredoxin-related protein from Desulfitobacterium hafniense DCB
To be Published
|
|
3MT1
 
 | | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti | | Descriptor: | Putative carboxynorspermidine decarboxylase protein, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti
To be Published
|
|
3GMI
 
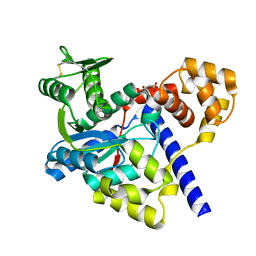 | |
3BWL
 
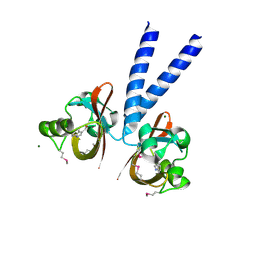 | | Crystal structure of PAS domain of HTR-like protein from Haloarcula marismortui | | Descriptor: | 1H-INDOLE-3-CARBALDEHYDE, MAGNESIUM ION, Sensor protein | | Authors: | Osipiuk, J, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-ray crystal structure of PAS domain of HTR-like protein from Haloarcula marismortui.
To be Published
|
|
3M0Z
 
 | | Crystal structure of putative aldolase from Klebsiella pneumoniae. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Chang, C, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of putative aldolase from Klebsiella pneumoniae.
To be Published
|
|
3BUU
 
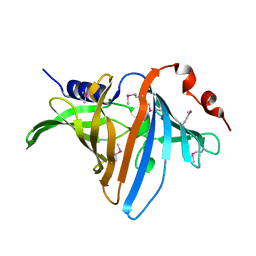 | |
3N2Q
 
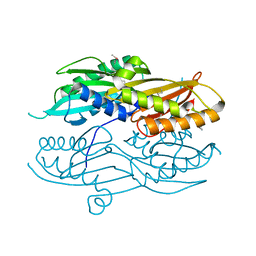 | |
3N70
 
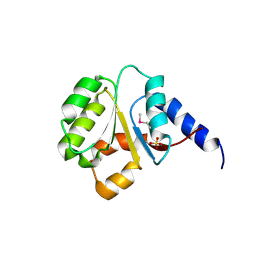 | | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A | | Descriptor: | SULFATE ION, Transport activator | | Authors: | Stein, A.J, Mulligan, R, Volkart, L, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the P-loop NTPase domain of the Sigma-54 transport activator from E. coli to 2.8A
To be Published
|
|
3GQS
 
 | | Crystal structure of the FHA domain of CT664 protein from Chlamydia trachomatis | | Descriptor: | Adenylate cyclase-like protein, PHOSPHATE ION | | Authors: | Majorek, K.A, Cymborowski, M, Chruszcz, M, Evdokimova, E, Egorova, O, Di Leo, R, Zimmerman, M.D, Savchenko, A, Joachimiak, A, Edwards, A.M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the FHA domain of CT664 protein from Chlamydia trachomatis
To be Published
|
|
3H92
 
 | |
3MCZ
 
 | |
3HA9
 
 | | The 1.7A Crystal Structure of a Thioredoxin-like Protein from Aeropyrum pernix | | Descriptor: | uncharacterized Thioredoxin-like protein | | Authors: | Stein, A.J, Cuff, M.E, Sather, A, Hendricks, R, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-01 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7A Crystal Structure of a Thioredoxin-like Protein from Aeropyrum pernix
To be Published
|
|
3B48
 
 | |
3HB7
 
 | | The Crystal Structure of an Isochorismatase-like Hydrolase from Alkaliphilus metalliredigens to 2.3A | | Descriptor: | AMMONIUM ION, Isochorismatase hydrolase, SODIUM ION | | Authors: | Stein, A.J, Xu, X, Cui, H, Ng, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-04 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of an Isochorismatase-like Hydrolase from Alkaliphilus metalliredigens to 2.3A
To be Published
|
|
3BJB
 
 | | Crystal structure of a TetR transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | Probable transcriptional regulator, TetR family protein, SULFATE ION | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-03 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of a TetR transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
3IUV
 
 | | The structure of a member of TetR family (SCO1917) from Streptomyces coelicolor A3 | | Descriptor: | uncharacterized TetR family protein | | Authors: | Tan, K, Cuff, M, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | The structure of a member of TetR family (SCO1917) from Streptomyces coelicolor A3
To be Published
|
|
3B7H
 
 | |
3H2Z
 
 | |
3B4S
 
 | |
3GYK
 
 | | The crystal structure of a thioredoxin-like oxidoreductase from Silicibacter pomeroyi DSS-3 | | Descriptor: | 1,2-ETHANEDIOL, 27kDa outer membrane protein, SULFATE ION | | Authors: | Fan, Y, Marshall, N, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-03 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The crystal structure of a thioredoxin-like oxidoreductase from Silicibacter pomeroyi DSS-3
To be Published
|
|
3MTJ
 
 | | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A | | Descriptor: | Homoserine dehydrogenase, SULFATE ION | | Authors: | Stein, A.J, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A
To be Published
|
|
3C8G
 
 | | Crystal structure of a possible transciptional regulator YggD from Shigella flexneri 2a str. 2457T | | Descriptor: | ACETATE ION, Putative transcriptional regulator | | Authors: | Tan, K, Borovilos, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-12 | | Release date: | 2008-02-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
