1K6D
 
 | | CRYSTAL STRUCTURE OF ACETATE COA-TRANSFERASE ALPHA SUBUNIT | | Descriptor: | ACETATE COA-TRANSFERASE ALPHA SUBUNIT, MAGNESIUM ION | | Authors: | Korolev, S, Koroleva, O, Petterson, K, Collart, F, Dementieva, I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-15 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autotracing of Escherichia coli acetate CoA-transferase alpha-subunit structure using 3.4 A MAD and 1.9 A native data.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KAF
 
 | | DNA Binding Domain Of The Phage T4 Transcription Factor MotA (AA105-211) | | Descriptor: | Transcription regulatory protein MOTA | | Authors: | Li, N, Sickmier, E.A, Zhang, R, Joachimiak, A, White, S.W. | | Deposit date: | 2001-11-01 | | Release date: | 2001-11-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The MotA transcription factor from bacteriophage T4 contains a novel DNA-binding domain: the 'double wing' motif.
Mol.Microbiol., 43, 2002
|
|
1I60
 
 | | Structural genomics, IOLI protein | | Descriptor: | IOLI PROTEIN | | Authors: | Zhang, R, Dementieva, I, Collart, F, Quaite-Randall, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-03-01 | | Release date: | 2002-03-13 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Bacillus subtilis ioli shows endonuclase IV fold with altered Zn binding.
Proteins, 48, 2002
|
|
1I6N
 
 | | 1.8 A Crystal structure of IOLI protein with a binding zinc atom | | Descriptor: | IOLI PROTEIN, ZINC ION | | Authors: | Zhang, R.G, Dementiva, I, Collart, F, Quaite-Randall, E, Joachimiak, A, Alkire, R, Maltsev, N, Korolev, O, Dieckman, L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-03-02 | | Release date: | 2002-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus subtilis ioli shows endonuclase IV fold with altered Zn binding.
Proteins, 48, 2002
|
|
1NJH
 
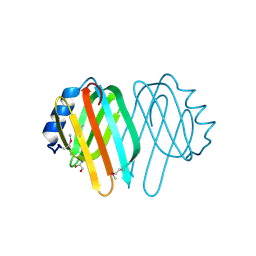 | |
6AWA
 
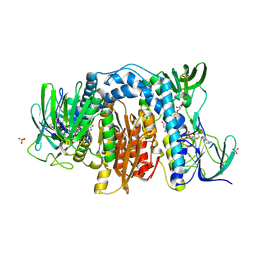 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate.
To Be Published
|
|
6AZI
 
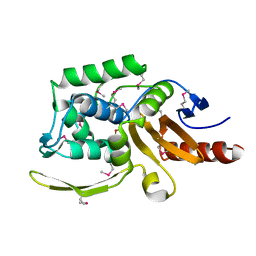 | | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid | | Descriptor: | BORATE ION, D-alanyl-D-alanine endopeptidase | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-11 | | Release date: | 2017-10-04 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid.
To be Published
|
|
6AZY
 
 | | Crystal structure of Hsp104 R328M/R757M mutant from Calcarisporiella thermophila | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein Hsp104 | | Authors: | Michalska, K, Bigelow, L, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-09-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
4HTN
 
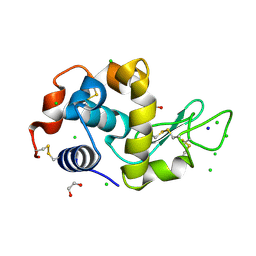 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 1.32 x 10e+12 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.A, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1JV2
 
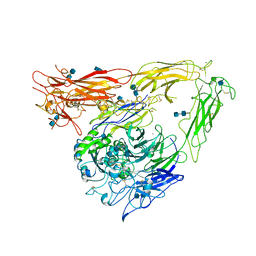 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN ALPHAVBETA3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.P, Stehle, T, Diefenbach, B, Zhang, R, Dunker, R, Scott, D, Joachimiak, A, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3.
Science, 294, 2001
|
|
6B8D
 
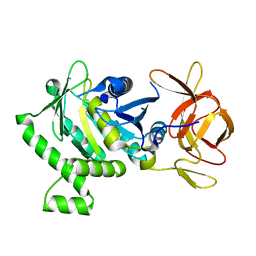 | | 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-06 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae.
To Be Published
|
|
6BBX
 
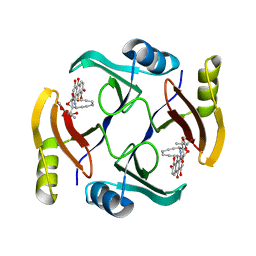 | | Crystal structure of TnmS3 in complex with TNM C | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2R,3R)-2,3-dihydroxy-3-[(1aS,11S,11aR,14Z,18R)-3,7,8,18-tetrahydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]butanoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-19 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
3B4S
 
 | |
4HTK
 
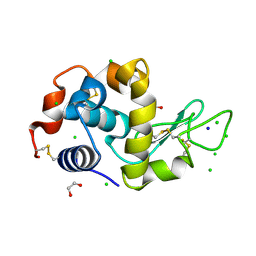 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 2.17 x 10e+12 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.A, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2IBD
 
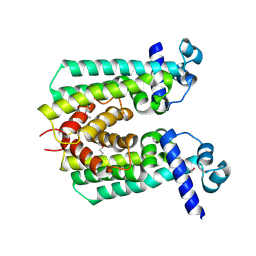 | | Crystal structure of Probable transcriptional regulatory protein RHA5900 | | Descriptor: | MAGNESIUM ION, Possible transcriptional regulator | | Authors: | Chang, C, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-11 | | Release date: | 2006-10-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Probable transcriptional regulatory protein RHA5900
To be Published
|
|
3B49
 
 | |
3B85
 
 | |
3B4Q
 
 | |
3B79
 
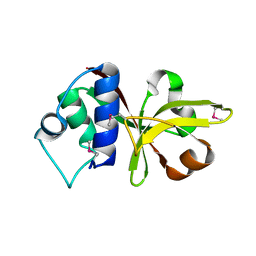 | |
1NEZ
 
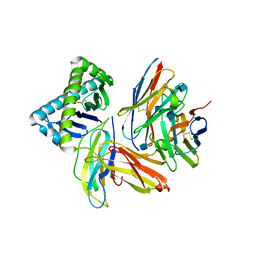 | | The Crystal Structure of a TL/CD8aa Complex at 2.1A resolution:Implications for Memory T cell Generation, Co-receptor Preference and Affinity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, Y, Xiong, Y, Naidenko, O.V, Liu, J.H, Zhang, R, Joachimiak, A, Kronenberg, M, Cheroutre, H, Reinherz, E.L, Wang, J.H. | | Deposit date: | 2002-12-12 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a TL/CD8alphaalpha Complex at 2.1 A resolution: Implications for modulation of T cell activation and memory
Immunity, 18, 2003
|
|
3BJN
 
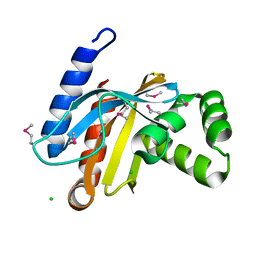 | | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae, targeted domain 79-240 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, putative | | Authors: | Chang, C, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae.
To be Published
|
|
1NG6
 
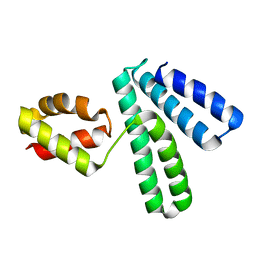 | | Structure of Cytosolic Protein of Unknown Function YqeY from Bacillus subtilis | | Descriptor: | Hypothetical protein yqeY | | Authors: | Zhang, R, Dementiva, I, Vinokour, E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-16 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4A crystal structure of hypothetical cytosolic protein
YQEY
To be Published
|
|
6B7J
 
 | |
6BZ0
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD. | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD.
To Be Published
|
|
1NSL
 
 | | Crystal structure of Probable acetyltransferase | | Descriptor: | CHLORIDE ION, Probable acetyltransferase | | Authors: | Brunzelle, J.S, Korolev, S.V, Wu, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YdaF protein: A putative ribosomal N-acetyltransferase
Proteins, 57, 2004
|
|
