8RYZ
 
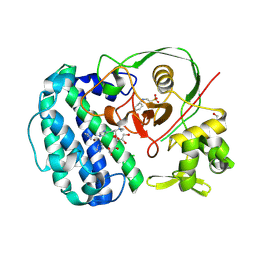 | | Structures of selenoneine synthase SenA from Variovorax paradoxus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, IMIDAZOLE, ... | | Authors: | Ma, Y.Y, Gao, Y, Xu, S.H. | | Deposit date: | 2024-02-11 | | Release date: | 2024-09-11 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structures of SenB and SenA enzymes from Variovorax paradoxus provide insights into carbon-selenium bond formation in selenoneine biosynthesis.
Heliyon, 10, 2024
|
|
7S1M
 
 | |
7S3I
 
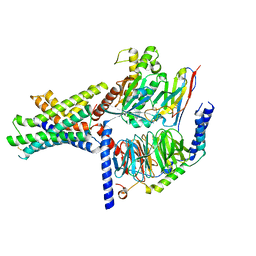 | | Ex4-D-Ala bound to the glucagon-like peptide-1 receptor/g protein complex (conformer 2) | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Belousoff, M.J, Piper, S.J, Danev, R. | | Deposit date: | 2021-09-07 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural and functional diversity among agonist-bound states of the GLP-1 receptor.
Nat.Chem.Biol., 18, 2022
|
|
9FI8
 
 | | SSU(head) structure derived from the SSU sample of the mitoribosome from T. gondii. | | Descriptor: | 30S ribosomal protein S5, putative, AP2 domain transcription factor AP2IX-6, ... | | Authors: | Rocha, R.E.O, Barua, S, Boissier, F, Nguyen, T.T, Hashem, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Apicomplexan mitoribosome from highly fragmented rRNAs to a functional machine.
Nat Commun, 15, 2024
|
|
9G6K
 
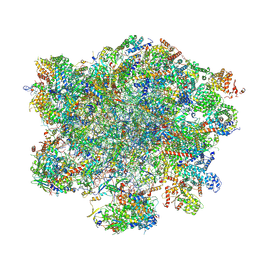 | | LSU structure derived from the LSU sample of the mitoribosome from T. gondii. | | Descriptor: | AMP-dependent synthetase/ligase domain-containing protein, AP2 domain transcription factor AP2IV-1, AP2 domain transcription factor AP2VIIb-2, ... | | Authors: | Rocha, R.E.O, Barua, S, Boissier, F, Nguyen, T.T, Hashem, Y. | | Deposit date: | 2024-07-18 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Apicomplexan mitoribosome from highly fragmented rRNAs to a functional machine.
Nat Commun, 15, 2024
|
|
9FIA
 
 | | SSU(body) structure derived from the SSU sample of the mitoribosome from T. gondii. | | Descriptor: | 30S ribosomal protein S12, putative, 30S ribosomal protein S5, ... | | Authors: | Rocha, R.E.O, Barua, S, Boissier, F, Nguyen, T.T, Hashem, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Apicomplexan mitoribosome from highly fragmented rRNAs to a functional machine.
Nat Commun, 15, 2024
|
|
8JZS
 
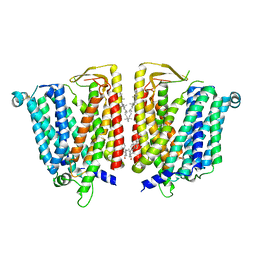 | | Outward-facing SLC15A4 dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, SLC15A4-TSLAA/EGPF tag protein | | Authors: | Zhang, S.S, Chen, X.D, Xie, M. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-27 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for recruitment of TASL by SLC15A4 in human endolysosomal TLR signaling.
Nat Commun, 14, 2023
|
|
8JZU
 
 | | SLC15A4_TASL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SLC15A4-TSLAA/EGPF tag protein, TLR adapter-Green fluorescent protein | | Authors: | Zhang, S.S, Chen, X.D, Xie, M. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-27 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for recruitment of TASL by SLC15A4 in human endolysosomal TLR signaling.
Nat Commun, 14, 2023
|
|
8JZR
 
 | | Outward_facing SLC15A4 monomer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SLC15A4-ALFA tag-SLC15A4-twin-strep tag fusion protein | | Authors: | Zhang, S.S, Chen, X.D, Xie, M. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-27 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for recruitment of TASL by SLC15A4 in human endolysosomal TLR signaling.
Nat Commun, 14, 2023
|
|
5NK0
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1j | | Descriptor: | 2-[[3-[(3-azanyl-2,2-dimethyl-propyl)carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NK4
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2c | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3-[[(4~{R})-3,3-bis(fluoranyl)piperidin-4-yl]carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKB
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 4a | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[(3,5-dimorpholin-4-ylphenyl)amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKH
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 3e | | Descriptor: | 2-[[3-(2-aminophenyl)-5-(piperidin-4-ylcarbamoyl)phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKE
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 3a | | Descriptor: | 2-[[3-bromanyl-5-(piperidin-4-ylcarbamoyl)phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NK3
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1l | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[[(3~{S})-pyrrolidin-3-yl]carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKC
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2h | | Descriptor: | (3~{S})-1-[3-[[5-[(2-chloranyl-6-methyl-phenyl)carbamoyl]-1,3-thiazol-2-yl]amino]phenyl]carbonylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
1QAJ
 
 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3'), REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-03-18 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain.
J.Mol.Biol., 296, 2000
|
|
1QAI
 
 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3'), MERCURY (II) ION, REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain.
J.Mol.Biol., 296, 2000
|
|
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8KHC
 
 | | SARS-CoV-2 Omicron spike in complex with 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
5NK2
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2b | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[[(3~{R},4~{R})-3-fluoranylpiperidin-4-yl]carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKA
 
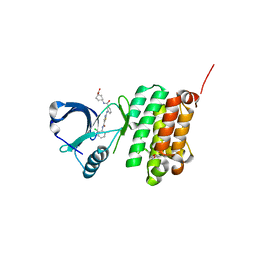 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2g | | Descriptor: | 1,2-ETHANEDIOL, 4-[[3-[[5-[(2-chloranyl-6-methyl-phenyl)carbamoyl]-1,3-thiazol-2-yl]amino]phenyl]carbonylamino]cyclohexane-1-carboxylic acid, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
8XIJ
 
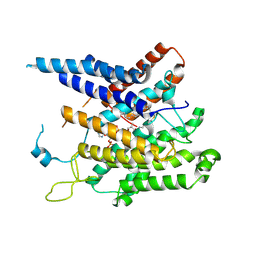 | | Structure of acyltransferase GWT1 bound to palmitoyl-CoA | | Descriptor: | GPI-anchored wall transfer protein 1, Palmitoyl-CoA | | Authors: | Dai, X.L, Li, J.L, Liu, X.Z, Deng, D, Yan, C.Y, Wang, X. | | Deposit date: | 2023-12-19 | | Release date: | 2024-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into the inhibition mechanism of fungal GWT1 by manogepix.
Nat Commun, 15, 2024
|
|
8XIK
 
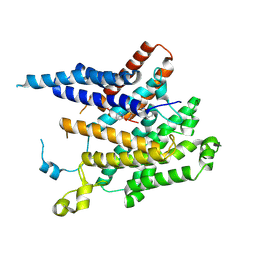 | | Structure of acyltransferase GWT1 in complex with manogepix(APX001A) | | Descriptor: | 3-[3-[[4-(pyridin-2-yloxymethyl)phenyl]methyl]-1,2-oxazol-5-yl]pyridin-2-amine, GPI-anchored wall transfer protein 1 | | Authors: | Dai, X.L, Li, J.L, Liu, X.Z, Deng, D, Wang, X. | | Deposit date: | 2023-12-19 | | Release date: | 2024-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural insights into the inhibition mechanism of fungal GWT1 by manogepix.
Nat Commun, 15, 2024
|
|
5NJZ
 
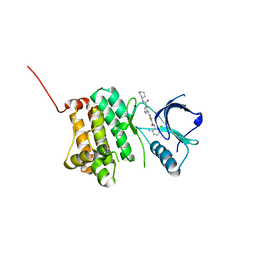 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1g | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-(piperidin-4-ylcarbamoyl)phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
