6ACB
 
 | | Crystal structure of PDE5 in complex with inhibitor LW1805 | | Descriptor: | 3-[(2H-1,3-benzodioxol-5-yl)methyl]-8-fluoro-6-{[2-(4-methylpiperazin-1-yl)ethyl]amino}-1-(1,3-thiazol-2-yl)[1]benzopyrano[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2018-07-26 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Optimization of Chromeno[2,3- c]pyrrol-9(2 H)-ones as Highly Potent, Selective, and Orally Bioavailable PDE5 Inhibitors: Structure-Activity Relationship, X-ray Crystal Structure, and Pharmacodynamic Effect on Pulmonary Arterial Hypertension.
J. Med. Chem., 61, 2018
|
|
4DNV
 
 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
7XBK
 
 | | Structure and mechanism of a mitochondrial AAA+ disaggregase CLPB | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Isoform 2 of Caseinolytic peptidase B protein homolog, MAGNESIUM ION, ... | | Authors: | Wu, D, Liu, Y, Dai, Y, Wang, G, Lu, G, Chen, Y, Li, N, Lin, J, Gao, N. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
5SY5
 
 | | Crystal Structure of the Heterodimeric NPAS1-ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Neuronal PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
5SY7
 
 | | Crystal Structure of the Heterodimeric NPAS3-ARNT Complex with HRE DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
8JSX
 
 | | VMAT2 complex with noradrenaline in lumen-facing state | | Descriptor: | Noradrenaline, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-20 | | Release date: | 2024-05-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
8JTB
 
 | | VMAT2 complex with dopamine | | Descriptor: | L-DOPAMINE, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2024-05-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
8JT5
 
 | | VMAT2 complex with histamine | | Descriptor: | HISTAMINE, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2024-05-08 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
8JSW
 
 | | Human VMAT2 complex with serotonin | | Descriptor: | SEROTONIN, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-29 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
8JTC
 
 | | Human VMAT2 complex with reserpine | | Descriptor: | Synaptic vesicular amine transporter, reserpine | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
8JT9
 
 | | Human VMAT2 complex with ketanserin | | Descriptor: | 3-[2-[4-(4-fluorophenyl)carbonylpiperidin-1-yl]ethyl]-1~{H}-quinazoline-2,4-dione, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
8JTA
 
 | | Human VMAT2 complex with tetrabenazine | | Descriptor: | Synaptic vesicular amine transporter, tetrabenazine | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
8XXS
 
 | | Crystal structure of PDE4D catalytic domain complexed with L11 | | Descriptor: | 2-[4-[bis(fluoranyl)methoxy]-3-(cyclopropylmethoxy)phenyl]-1-benzofuran-6-ol, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.10000944 Å) | | Cite: | 5-hydroxymethylcytosine features of portal venous blood predict metachronous liver metastases of colorectal cancer and reveal phosphodiesterase 4 as a therapeutic target.
Clin Transl Med, 15, 2025
|
|
5H2Z
 
 | |
8XOA
 
 | | VMAT2 complex with MPP+ | | Descriptor: | 1-methyl-4-phenylpyridin-1-ium, Synaptic vesicular amine transporter,Synaptic vesicular amine transporter,transporterA | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-12-31 | | Release date: | 2024-05-08 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
8XO9
 
 | | VMAT2 complex with noradrenaline in cytosol-facing state | | Descriptor: | Noradrenaline, Synaptic vesicular amine transporter,Synaptic vesicular amine transporter,transporter A | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-12-31 | | Release date: | 2024-05-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
8XOB
 
 | | VMAT2 protonated state | | Descriptor: | Synaptic vesicular amine transporter,transporter B | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-12-31 | | Release date: | 2024-05-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
6IKM
 
 | | Crystal structure of SpuE-Spermidine in complex with ScFv5 | | Descriptor: | Polyamine transport protein, SPERMIDINE, SULFATE ION, ... | | Authors: | Wu, D, Sun, X. | | Deposit date: | 2018-10-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | A Potent Anti-SpuE Antibody Allosterically Inhibits Type III Secretion System and Attenuates Virulence of Pseudomonas Aeruginosa.
J.Mol.Biol., 431, 2019
|
|
7XC5
 
 | | Crystal structure of the ANK domain of CLPB | | Descriptor: | Isoform 2 of Caseinolytic peptidase B protein homolog | | Authors: | Liu, Y, Wu, D, Lu, G, Gao, N, Lin, J. | | Deposit date: | 2022-03-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
3D04
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with sakuranetin | | Descriptor: | (2S)-5-hydroxy-2-(4-hydroxyphenyl)-7-methoxy-2,3-dihydro-4H-chromen-4-one, (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, ... | | Authors: | Zhang, L, Kong, Y, Wu, D, Shen, X, Jiang, H. | | Deposit date: | 2008-05-01 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay
Protein Sci., 17, 2008
|
|
9LO7
 
 | | The crystal structure of PDE4D with 2317b | | Descriptor: | 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huag, Y.-Y, Wu, D, Luo, H.-B. | | Deposit date: | 2025-01-22 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.20006251 Å) | | Cite: | Structure-Based Optimization of Moracin M as Potent and Selective PDE4 Inhibitors with Antipsoriasis Effects.
J.Med.Chem., 68, 2025
|
|
6KUW
 
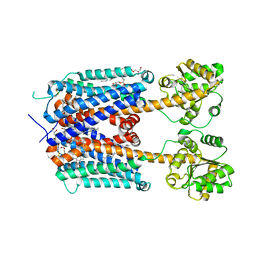 | | Crystal structure of human alpha2C adrenergic G protein-coupled receptor. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8~{a}~{R},12~{a}~{S},13~{a}~{R})-12-ethylsulfonyl-3-methoxy-5,6,8,8~{a},9,10,11,12~{a},13,13~{a}-decahydroisoquinolino[2,1-g][1,6]naphthyridine, Alpha-2C adrenergic receptor, ... | | Authors: | Chen, X.Y, Wu, L.J, Wu, D, Zhong, G.S. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human alpha2C adrenergic G protein-coupled receptor.
To Be Published
|
|
8QCS
 
 | |
8QCT
 
 | |
