2KZN
 
 | | Solution NMR Structure of Peptide methionine sulfoxide reductase msrB from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR10 | | Descriptor: | Peptide methionine sulfoxide reductase msrB | | Authors: | Ertekin, A, Maglaqui, M, Janjua, H, Cooper, B, Ciccosanti, C, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J, Lee, H, Aramini, J.M, Rossi, P, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LOK
 
 | | Solution NMR Structure of the uncharacterized protein from gene locus VNG_0733H of Halobacterium salinarium, Northeast Structural Genomics Consortium Target HsR50 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Lange, O.F, Lee, H, Hamilton, K, Ciccosanti, C, Buchwald, W.A, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-24 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2N44
 
 | | EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N45
 
 | | EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data with a second set of RDC data simulated for an alternative alignment tensor. Northeast Structural Genomics Consortium target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
7SKN
 
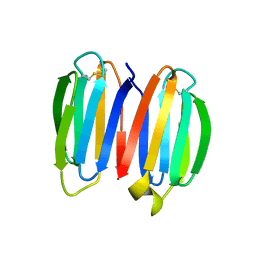 | |
7SKP
 
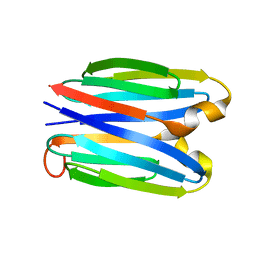 | |
7SKO
 
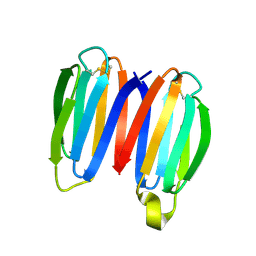 | | De novo synthetic protein DIG8-CC (orthogonal space group) | | Descriptor: | De novo synthetic protein DIG8-CC, MAGNESIUM ION | | Authors: | Mendes, S.R, Eckhard, U, Marcos, E, Gomis-Ruth, F.X. | | Deposit date: | 2021-10-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | De novo design of immunoglobulin-like domains
Nat Commun, 13, 2022
|
|
2N47
 
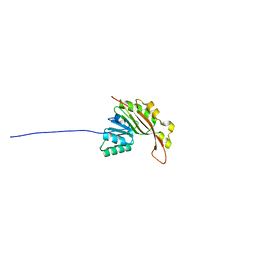 | | EC-NMR Structure of Synechocystis sp. PCC 6803 Slr1183 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
