2CG4
 
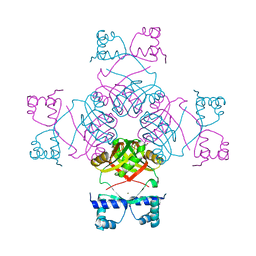 | | Structure of E.coli AsnC | | Descriptor: | ASPARAGINE, MAGNESIUM ION, REGULATORY PROTEIN ASNC | | Authors: | Thaw, P, Sedelnikova, S.E, Muranova, T, Wiese, S, Ayora, S, Alonso, J.C, Brinkman, A.B, Akerboom, J, van der Oost, J, Rafferty, J.B. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into Gene Transcriptional Regulation and Effector Binding by the Lrp/Asnc Family.
Nucleic Acids Res., 34, 2006
|
|
1Q8R
 
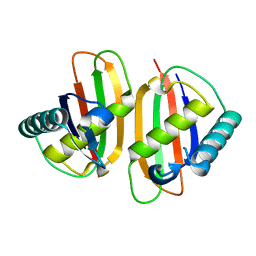 | | Structure of E.coli RusA Holliday junction resolvase | | Descriptor: | Crossover junction endodeoxyribonuclease rusA | | Authors: | Rafferty, J.B, Bolt, E.L, Muranova, T.A, Sedelnikova, S.E, Leonard, P, Pasquo, A, Baker, P.J, Rice, D.W, Sharples, G.J, Lloyd, R.G. | | Deposit date: | 2003-08-22 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | The structure of Escherichia coli RusA endonuclease reveals a new Holliday junction DNA binding fold
Structure, 11, 2003
|
|
1L5J
 
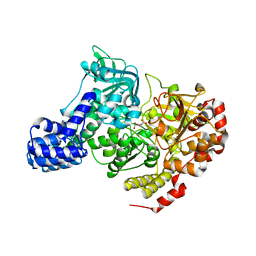 | | CRYSTAL STRUCTURE OF E. COLI ACONITASE B. | | Descriptor: | ACONITATE ION, Aconitate hydratase 2, FE3-S4 CLUSTER | | Authors: | Williams, C.H, Stillman, T.J, Barynin, V.V, Sedelnikova, S.E, Tang, Y, Green, J, Guest, J.R, Artymiuk, P.J. | | Deposit date: | 2002-03-07 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | E. coli aconitase B structure reveals a HEAT-like domain with implications for protein-protein recognition.
Nat.Struct.Biol., 9, 2002
|
|
1NO1
 
 | | Structure of truncated variant of B.subtilis SPP1 phage G39P helicase loader/inhibitor protein | | Descriptor: | replisome organizer | | Authors: | Bailey, S, Sedelnikova, S.E, Mesa, P, Ayora, S, Waltho, J.P, Ashcroft, A.E, Baron, A.J, Alonso, J.C, Rafferty, J.B. | | Deposit date: | 2003-01-15 | | Release date: | 2003-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of Bacillus subtilis SPP1 phage helicase loader protein G39P
J.Biol.Chem., 278, 2003
|
|
1LEH
 
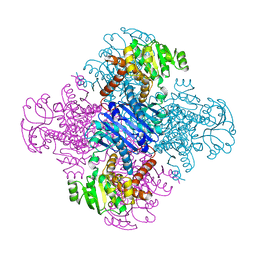 | | LEUCINE DEHYDROGENASE FROM BACILLUS SPHAERICUS | | Descriptor: | LEUCINE DEHYDROGENASE | | Authors: | Baker, P.J, Turnbull, A.P, Sedelnikova, S.E, Stillman, T.J, Rice, D.W. | | Deposit date: | 1995-06-09 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A role for quaternary structure in the substrate specificity of leucine dehydrogenase.
Structure, 3, 1995
|
|
1IGW
 
 | | Crystal Structure of the Isocitrate Lyase from the A219C mutant of Escherichia coli | | Descriptor: | Isocitrate lyase, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Britton, K.L, Abeysinghe, I.S.B, Baker, P.J, Barynin, V, Diehl, P, Langridge, S.J, McFadden, B.A, Sedelnikova, S.E, Stillman, T.J, Weeradechapon, K, Rice, D.W. | | Deposit date: | 2001-04-18 | | Release date: | 2001-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure and domain organization of Escherichia coli isocitrate lyase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1BVU
 
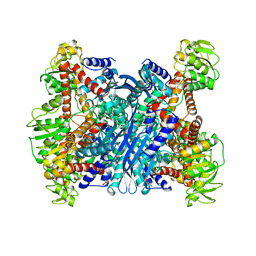 | | GLUTAMATE DEHYDROGENASE FROM THERMOCOCCUS LITORALIS | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Baker, P.J, Britton, K.L, Yip, K.S, Stillman, T.J, Rice, D.W. | | Deposit date: | 1999-07-20 | | Release date: | 1999-09-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of the glutamate dehydrogenase from the hyperthermophile Thermococcus litoralis and its comparison with that from Pyrococcus furiosus
J.Mol.Biol., 293, 1999
|
|
6EZM
 
 | | Imidazoleglycerol-phosphate dehydratase from Saccharomyces cerevisiae | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION, [(2R)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]phosphonic acid | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EZJ
 
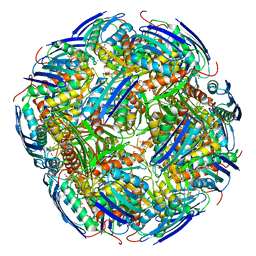 | | Imidazoleglycerol-phosphate dehydratase | | Descriptor: | Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, MANGANESE (II) ION, ... | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5LBM
 
 | | The asymmetric tetrameric structure of the formaldehyde sensing transcriptional repressor FrmR from Escherichia coli | | Descriptor: | FORMYL GROUP, Transcriptional repressor FrmR | | Authors: | Bisson, C, Baker, P.J, Green, J, Chivers, P.T. | | Deposit date: | 2016-06-16 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The mechanism of a formaldehyde-sensing transcriptional regulator.
Sci Rep, 6, 2016
|
|
4K1P
 
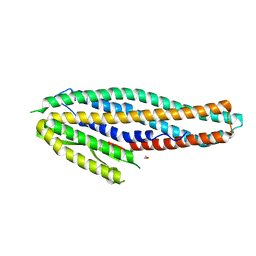 | | Structure of the NheA component of the Nhe toxin from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, NheA, SULFATE ION | | Authors: | Ganash, M, Phung, D, Artymiuk, P.J. | | Deposit date: | 2013-04-05 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the NheA Component of the Nhe Toxin from Bacillus cereus: Implications for Function.
Plos One, 8, 2013
|
|
4DAP
 
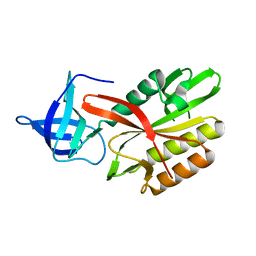 | | The structure of Escherichia coli SfsA | | Descriptor: | SODIUM ION, Sugar fermentation stimulation protein A | | Authors: | Baker, P.J, Allen, F.L. | | Deposit date: | 2012-01-13 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of SfsA and its DNA complex; A DNA/RNA nuclease with a novel domain combination
To be Published
|
|
4DAV
 
 | | The structure of Pyrococcus Furiosus SfsA in complex with DNA | | Descriptor: | 5'-D(*CP*GP*CP*TP*GP*TP*CP*TP*CP*GP*CP*T)-3', Sugar fermentation stimulation protein homolog | | Authors: | Baker, P.J, Allen, F.L. | | Deposit date: | 2012-01-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of SfsA and its DNA complex; A DNA/RNA nuclease with a novel domain combination
To be Published
|
|
6ZZB
 
 | |
6ZZH
 
 | |
7A27
 
 | |
6ZZ5
 
 | | Structure of soluble SmhB of the tripartite alpha-pore forming toxin, Smh, from Serratia marcescens. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, SmhB | | Authors: | Churchill-Angus, A.M, Baker, P.J. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Characterisation of a tripartite alpha-pore forming toxin from Serratia marcescens
Sci Rep, 11, 2021
|
|
7A26
 
 | |
7A0G
 
 | |
1CUK
 
 | |
4LUL
 
 | |
4LUK
 
 | |
4LTA
 
 | |
4LUM
 
 | |
6FWH
 
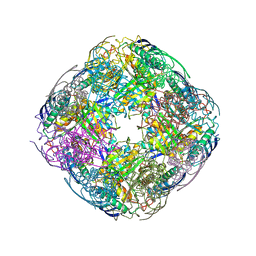 | | Acanthamoeba IGPD in complex with R-C348 to 1.7A resolution | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Roberts, C.W, Bisson, C, Baker, P.J. | | Deposit date: | 2018-03-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional studies of histidine biosynthesis in Acanthamoeba spp. demonstrates a novel molecular arrangement and target for antimicrobials.
PLoS ONE, 13, 2018
|
|
