5GTH
 
 | | Native XFEL structure of photosystem II (dark dataset) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-08-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
4YOP
 
 | | CRYSTAL STRUCTURE OF HEN EGG-WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Nakane, T, Suzuki, M, Nango, E. | | Deposit date: | 2015-03-12 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Native sulfur/chlorine SAD phasing for serial femtosecond crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8W6X
 
 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Hiromoto, T, Tamada, T. | | Deposit date: | 2023-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | Cite: | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
5WS5
 
 | | Native XFEL structure of photosystem II (preflash dark dataset) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
5WS6
 
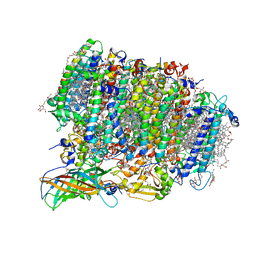 | | Native XFEL structure of Photosystem II (preflash two-flash dataset | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
1R77
 
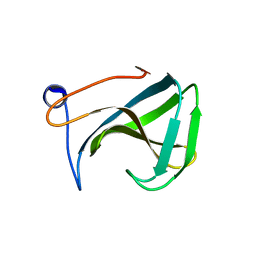 | | Crystal structure of the cell wall targeting domain of peptidylglycan hydrolase ALE-1 | | Descriptor: | Cell Wall Targeting Domain of Glycylglycine Endopeptidase ALE-1 | | Authors: | Lu, J.Z, Fujiwara, T, Komatsuzawa, H, Sugai, M, Sakon, J. | | Deposit date: | 2003-10-20 | | Release date: | 2005-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cell Wall-targeting Domain of Glycylglycine Endopeptidase Distinguishes among Peptidoglycan Cross-bridges.
J.Biol.Chem., 281, 2006
|
|
7DI8
 
 | |
3VYE
 
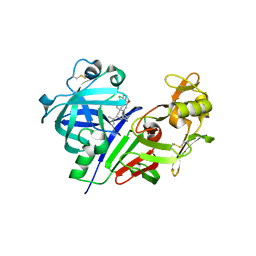 | | Human renin in complex with inhibitor 7 | | Descriptor: | (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]-N-(3-methylbutyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Takahashi, M, Matsui, Y, Hanzawa, H. | | Deposit date: | 2012-09-24 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and discovery of new (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]piperidine-3-carboxamides as potent renin inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3VYD
 
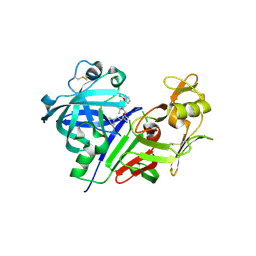 | | Human renin in complex with inhibitor 6 | | Descriptor: | (3S,5R)-5-{[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]methyl}-N-(3-methylbutyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2012-09-24 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Design and discovery of new (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]piperidine-3-carboxamides as potent renin inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3VYF
 
 | | Human renin in complex with inhibitor 9 | | Descriptor: | (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]-N-(2,6-dimethylheptan-4-yl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2012-09-24 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and discovery of new (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]piperidine-3-carboxamides as potent renin inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5GSY
 
 | | Kinesin-8 motor, KIF19A, in the nucleotide-free state complexed with GDP-taxol microtubule | | Descriptor: | Kinesin-like protein KIF19 | | Authors: | Morikawa, M, Nitta, R, Yajima, H, Shigematsu, H, Kikkawa, M, Hirokawa, N. | | Deposit date: | 2016-08-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Elife, 5, 2016
|
|
5GSZ
 
 | | Crystal Structure of the KIF19A Motor Domain Complexed with Mg-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF19, MAGNESIUM ION | | Authors: | Wang, D, Nitta, R, Hirokawa, N. | | Deposit date: | 2016-08-18 | | Release date: | 2016-09-28 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Elife, 5, 2016
|
|
2Z67
 
 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) selenium transferase (SepSecS) | | Descriptor: | O-phosphoseryl-tRNA(Sec) selenium transferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Araiso, Y, Ishitani, R, Pailouer, S, Oshikane, H, Domae, N, Soll, D, Nureki, O. | | Deposit date: | 2007-07-23 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formation.
Nucleic Acids Res., 36, 2008
|
|
3VW5
 
 | |
