3C9B
 
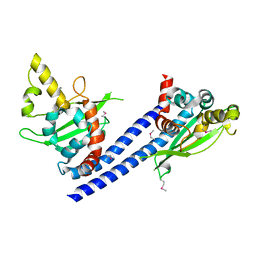 | | Crystal structure of SeMet Vps75 | | Descriptor: | Vacuolar protein sorting-associated protein 75 | | Authors: | Keck, J.L, Berndsen, C.E, Tsubota, T, Lindner, S.E, Lee, S, Holton, J.M, Kaufman, P.D, Denu, J.M. | | Deposit date: | 2008-02-15 | | Release date: | 2008-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75
Nat.Struct.Mol.Biol., 15, 2008
|
|
1J1B
 
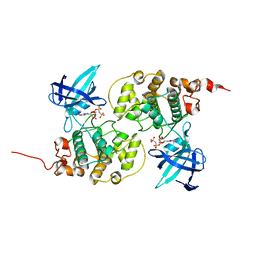 | | Binary complex structure of human tau protein kinase I with AMPPNP | | Descriptor: | Glycogen synthase kinase-3 beta, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Aoki, M, Yokota, T, Sugiura, I, Sasaki, C, Hasegawa, T, Okumura, C, Kohno, T, Sugio, S, Matsuzaki, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 beta.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2IDC
 
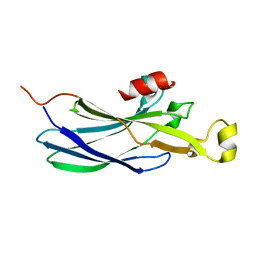 | | Structure of the Histone H3-Asf1 Chaperone Interaction | | Descriptor: | ANTI-SILENCING PROTEIN 1 AND HISTONE H3 CHIMERA | | Authors: | Antczak, A.J, Tsubota, T, Kaufman, P.D, Berger, J.M. | | Deposit date: | 2006-09-14 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics.
Bmc Struct.Biol., 6, 2006
|
|
1UER
 
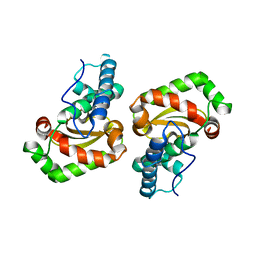 | | Crystal structure of Porphyromonas gingivalis SOD | | Descriptor: | FE (III) ION, superoxide dismutase | | Authors: | Yamakura, F, Sugio, S, Hiraoka, B.Y, Yokota, T, Ohmori, D. | | Deposit date: | 2003-05-20 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pronounced conversion of the metal-specific activity of superoxide dismutase from Porphyromonas gingivalis by the mutation of a single amino acid (Gly155Thr) located apart from the active site
Biochemistry, 42, 2003
|
|
1UES
 
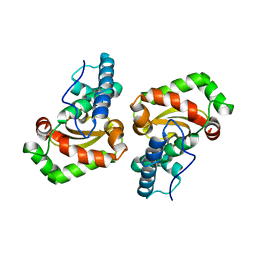 | | Crystal structure of Porphyromonas gingivalis SOD | | Descriptor: | MANGANESE (II) ION, superoxide dismutase | | Authors: | Yamakura, F, Sugio, S, Hiraoka, B.Y, Yokota, T, Ohmori, D. | | Deposit date: | 2003-05-20 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pronounced conversion of the metal-specific activity of superoxide dismutase from Porphyromonas gingivalis by the mutation of a single amino acid (Gly155Thr) located apart from the active site
Biochemistry, 42, 2003
|
|
1HZK
 
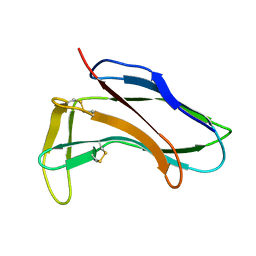 | |
1HZL
 
 | | SOLUTION STRUCTURES OF C-1027 APOPROTEIN AND ITS COMPLEX WITH THE AROMATIZED CHROMOPHORE | | Descriptor: | C-1027 APOPROTEIN, C-1027 AROMATIZED CHROMOPHORE | | Authors: | Tanaka, T, Fukuda-Ishisaka, S, Hirama, M, Otani, T. | | Deposit date: | 2001-01-25 | | Release date: | 2001-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of C-1027 apoprotein and its complex with the aromatized chromophore.
J.Mol.Biol., 309, 2001
|
|
8WQP
 
 | |
7VKA
 
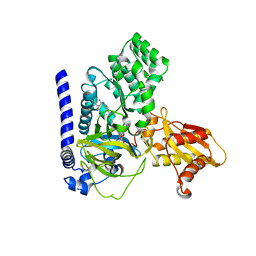 | | Crystal Structure of GH3.6 in complex with an inhibitor | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Indole-3-acetic acid-amido synthetase GH3.6, ... | | Authors: | Wang, N, Luo, M, Bao, H, Huang, H. | | Deposit date: | 2021-09-29 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Chemical genetic screening identifies nalacin as an inhibitor of GH3 amido synthetase for auxin conjugation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8IDS
 
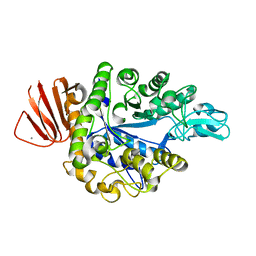 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258P in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
8IBK
 
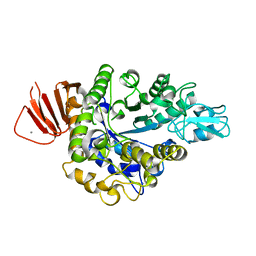 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258G in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
7C8L
 
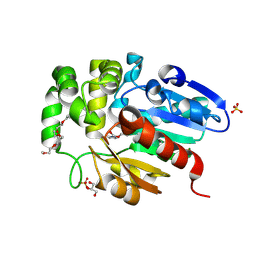 | | Hybrid designing of potent inhibitors of Striga strigolactone receptor ShHTL7 | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, GLYCEROL, Hyposensitive to light 7, ... | | Authors: | Shahul Hameed, U.F, Arold, S.T. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rational design of Striga hermonthica-specific seed germination inhibitors.
Plant Physiol., 188, 2022
|
|
4JPJ
 
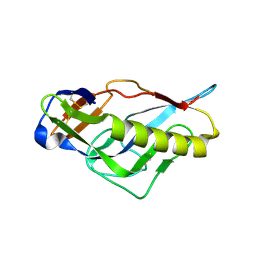 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT6 | | Authors: | Julien, J.-P, Jardine, J, Schief, W.R, Wilson, I.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational HIV immunogen design to target specific germline B cell receptors.
Science, 340, 2013
|
|
4JPI
 
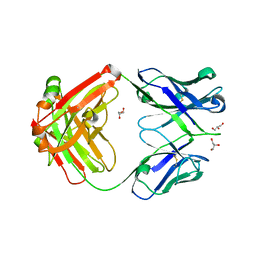 | | Crystal structure of a putative VRC01 germline precursor Fab | | Descriptor: | GLYCEROL, Putative VRC01 germline Fab heavy chain, Putative VRC01 germline Fab light chain | | Authors: | Julien, J.-P, Diwanji, D.C, Jardine, J, Schief, W.R, Wilson, I.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational HIV immunogen design to target specific germline B cell receptors.
Science, 340, 2013
|
|
4JPK
 
 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain eOD-GT6 in complex with a putative VRC01 germline precursor Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT6, ... | | Authors: | Julien, J.-P, Jardine, J, Schief, W.R, Wilson, I.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational HIV immunogen design to target specific germline B cell receptors.
Science, 340, 2013
|
|
6OL5
 
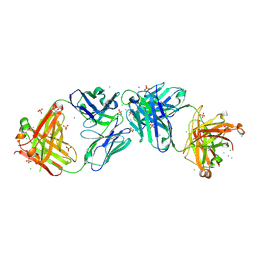 | |
6OL6
 
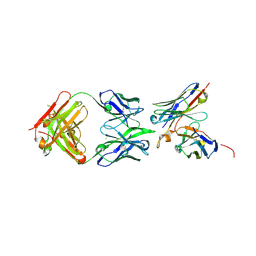 | | Structure of iglb12 scFv in complex with anti-idiotype ib2 Fab | | Descriptor: | CHLORIDE ION, ib2 Heavy Chain, ib2 Light Chain, ... | | Authors: | Weidle, C, Pancera, M, Gewe, M. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Detection and activation of HIV broadly neutralizing antibody precursor B cells using anti-idiotypes.
J.Exp.Med., 216, 2019
|
|
5WL2
 
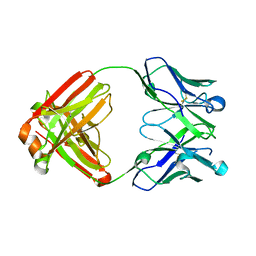 | | VH1-69 germline antibody with CDR H3 sequence of CR9114 | | Descriptor: | Germline-reverted light chain of CR9114, Heavy chain of VH1-69 germline antibody with CDR H3 sequence of CR9114 | | Authors: | Wilson, I.A, Lang, S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anti-idiotypic antibody K1-18 engages VH1-69 precursor and affinity-matured, anti-stem antibodies through mimicry of the HA stem.
To Be Published
|
|
5WKZ
 
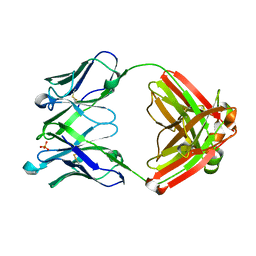 | | VH1-69 germline antibody predicted from CR6261 | | Descriptor: | Immunoglobulin heavy variable 1-69D,IgG H chain, Lambda-chain (AA -20 to 215), SULFATE ION | | Authors: | Lang, S, Lee, P.S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Anti-idiotypic antibody K1-18 engages VH1-69 precursor and affinity-matured, anti-stem antibodies through mimicry of the HA stem
To Be Published
|
|
1C8I
 
 | | BINDING MODE OF HYDROXYLAMINE TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROXYAMINE, ... | | Authors: | Wariishi, H, Nonaka, D, Johjima, T, Nakamura, N, Naruta, Y, Kubo, K, Fukuyama, K. | | Deposit date: | 2000-05-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct binding of hydroxylamine to the heme iron of Arthromyces ramosus peroxidase. Substrate analogue that inhibits compound I formation in a competetive manner.
J.Biol.Chem., 275, 2000
|
|
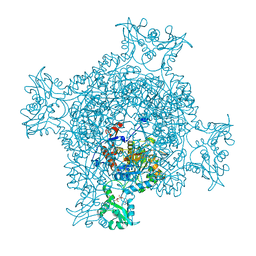
6JNF
 
 | | Cryo-EM structure of the translocator of the outer mitochondrial membrane | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Araiso, Y, Tsutsumi, A, Suzuki, J, Yunoki, K, Kawano, S, Kikkawa, M, Endo, T. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure of the mitochondrial import gate reveals distinct preprotein paths.
Nature, 575, 2019
|
|
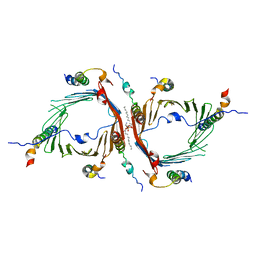
6JIG
 
 | | Crystal structure of GMP reductase C318A from Trypanosoma brucei in complex with guanosine 5'-monophosphate | | Descriptor: | GMP reductase, GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION | | Authors: | Mase, H, Imamura, A, Nishimura, S, Inui, T. | | Deposit date: | 2019-02-21 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Allosteric regulation accompanied by oligomeric state changes of Trypanosoma brucei GMP reductase through cystathionine-beta-synthase domain.
Nat Commun, 11, 2020
|
|
6JL8
 
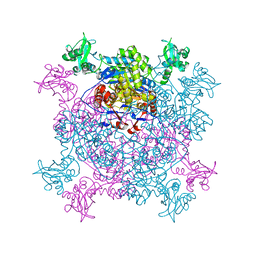 | |
6A1C
 
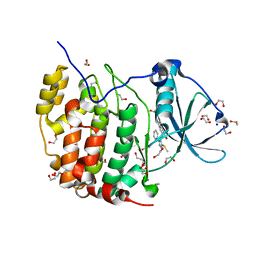 | | Crystal structure of the CK2a1-go289 complex | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-4-[(E)-(3-methylsulfanyl-5-phenyl-1,2,4-triazol-4-yl)iminomethyl]phenol, Casein kinase II subunit alpha, ... | | Authors: | Kinoshita, T, Tsuyuguchi, M. | | Deposit date: | 2018-06-07 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth.
Sci Adv, 5, 2019
|
|
6A9E
 
 | | Crystal structure of the N-terminal domain of Atg2 | | Descriptor: | Endolysin,Autophagy-related protein 2 | | Authors: | Osawa, T, Noda, N.N. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Atg2 mediates direct lipid transfer between membranes for autophagosome formation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
