8I40
 
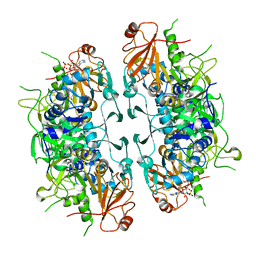 | | Crystal structure of ASCT from Trypanosoma brucei in complex with CoA. | | Descriptor: | ACETATE ION, CALCIUM ION, COENZYME A, ... | | Authors: | Mochizuki, K, Inaoka, D.K, Fukuda, K, Kurasawa, H, Iyoda, K, Nakai, U, Harada, S, Balogun, E.O, Mazet, M, Millerioux, Y, Bringaud, F, Boshart, M, Hirayama, K, Kita, K, Shiba, T. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of ligand complexes of ASCT from Trypanosoma brucei and molecular mechanism in comparison with mammalian SCOT.
To Be Published
|
|
5E8D
 
 | | Crystal structure of human epiregulin in complex with the Fab fragment of murine monoclonal antibody 9E5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Proepiregulin, ... | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
5AZ2
 
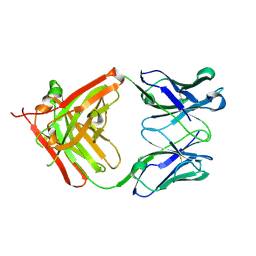 | | Crystal structure of the Fab fragment of 9E5, a murine monoclonal antibody specific for human epiregulin | | Descriptor: | anti-human epiregulin antibody 9E5 Fab heavy chain, anti-human epiregulin antibody 9E5 Fab light chain | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
4WRI
 
 | | Crystal structure of okadaic acid binding protein 2.1 | | Descriptor: | OKADAIC ACID, Okadaic acid binding protein 2-alpha | | Authors: | Ehara, H, Makino, M, Kodama, K, Ito, T, Sekine, S, Fukuzawa, S, Yokoyama, S, Tachibana, K. | | Deposit date: | 2014-10-24 | | Release date: | 2015-05-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Okadaic Acid Binding Protein 2.1: A Sponge Protein Implicated in Cytotoxin Accumulation
Chembiochem, 16, 2015
|
|
8XW5
 
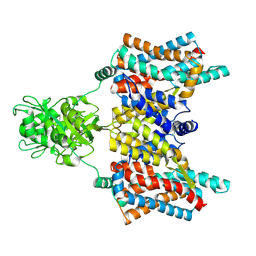 | | Cryo-EM structure of the aspartate:alanine antiporter AspT mutant L60C | | Descriptor: | Aspartate/alanine antiporter | | Authors: | Nanatani, K, Kanno, R, Kawabata, T, Watanabe, S, Hidaka, M, Yamanaka, T, Toda, K, Fujiki, T, Kunii, K, Miyamoto, A, Chiba, F, Ogasawara, S, Murata, T, Humbel, B.M, Inaba, K, Mitsuoka, K, Guan, L, Abe, K, Yamamoto, M, Koshiba, S. | | Deposit date: | 2024-01-16 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the aspartate:alanine antiporter AspT from Tetragenococcus halophilus
To Be Published
|
|
8Y1X
 
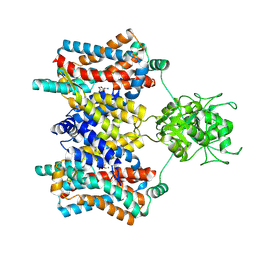 | | Cryo-EM structure of the aspartate:alanine antiporter AspT WT | | Descriptor: | ASPARTIC ACID, Aspartate/alanine antiporter | | Authors: | Nanatani, K, Kanno, R, Kawabata, T, Watanabe, S, Hidaka, M, Yamanaka, T, Toda, K, Fujiki, T, Kunii, K, Miyamoto, A, Chiba, F, Ogasawara, S, Murata, T, Humbel, B.M, Inaba, K, Mitsuoka, K, Guan, L, Abe, K, Yamamoto, M, Koshiba, S. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the aspartate:alanine antiporter AspT from Tetragenococcus halophilus
To Be Published
|
|
1GC2
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
1GC0
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
2RQ9
 
 | | Solution structure of human acidic fibroblast growth factor (aFGF) in the presence of a protein stabilizer NDSB-new | | Descriptor: | Heparin-binding growth factor 1 | | Authors: | Enomoto, M, Xiang, L, Ishii, T, Tochio, N, Hosoda, K, Inoue, Y, Nameki, N, Wakamatsu, K. | | Deposit date: | 2009-03-12 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human acidic fibroblast growth factor (aFGF) in the presence of a protein stabilizer NDSB-new
To be Published
|
|
4I86
 
 | | Crystal structure of PilZ domain of CeSA from cellulose synthesizing bacterium | | Descriptor: | Cellulose synthase 1 | | Authors: | Fujiwara, T, Komoda, K, Sakurai, N, Tanaka, I, Yao, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A
Biochem.Biophys.Res.Commun., 431, 2013
|
|
4P76
 
 | | Cellular response to a crystal-forming protein | | Descriptor: | Photoconvertible fluorescent protein, SODIUM ION | | Authors: | Tsutsui, H, Jinno, Y, Shoda, K, Tomita, A, Matsuda, M, Yamashita, E, Katayama, H, Nakagawa, A, Miyawaki, A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A diffraction-quality protein crystal processed as an autophagic cargo
Mol.Cell, 58, 2015
|
|
1JUD
 
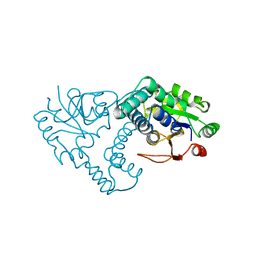 | | L-2-HALOACID DEHALOGENASE | | Descriptor: | L-2-HALOACID DEHALOGENASE | | Authors: | Hisano, T, Hata, Y, Fujii, T, Liu, J.-Q, Kurihara, T, Esaki, N, Soda, K. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of L-2-haloacid dehalogenase from Pseudomonas sp. YL. An alpha/beta hydrolase structure that is different from the alpha/beta hydrolase fold.
J.Biol.Chem., 271, 1996
|
|
8Z5L
 
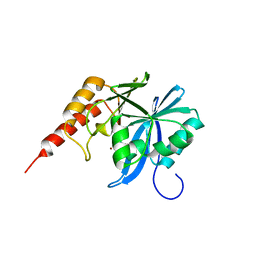 | | Crystal structure of metallo-beta-lactamse, IMP-1, complexed with a quinolinone-based inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-cyclohexylethyl-[3-(4-methylphenoxy)propyl]amino]phenyl]propanoic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Nimura, S, Guo, Y, Kondo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Development of Inhibitory Compounds for Metallo-beta-lactamase through Computational Design and Crystallographic Analysis.
Biochemistry, 63, 2024
|
|
6JKA
 
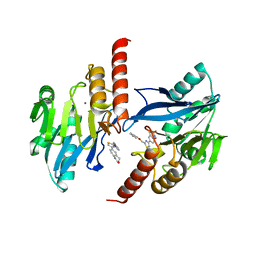 | | Crystal structure of metallo-beta-lactamse, IMP-1, in complex with a thiazole-bearing inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-(phenoxymethyl)-1,3-thiazol-4-yl]phenyl]propanoic acid, 6-[2-(phenoxymethyl)-1,3-thiazol-4-yl]-3,4-dihydro-1H-quinolin-2-one, Metallo-beta-lactamase type 2, ... | | Authors: | Kamo, T, Kuroda, K, Kondo, S, Hayashi, U, Fudo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Identification of the Inhibitory Compounds for Metallo-beta-lactamases and Structural Analysis of the Binding Modes.
Chem Pharm Bull (Tokyo), 69, 2021
|
|
6JKB
 
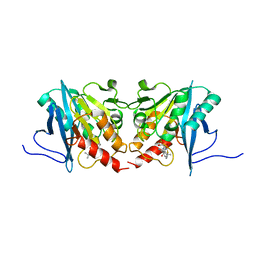 | | Crystal structure of metallo-beta-lactamse, NDM-1, in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Kondo, S, Hayashi, U, Fudo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Identification of the Inhibitory Compounds for Metallo-beta-lactamases and Structural Analysis of the Binding Modes.
Chem Pharm Bull (Tokyo), 69, 2021
|
|
1IOZ
 
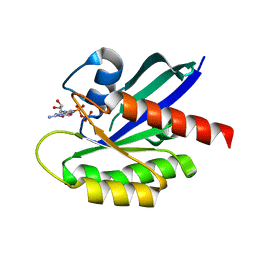 | | Crystal Structure of the C-HA-RAS Protein Prepared by the Cell-Free Synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Kigawa, T, Yamaguchi-Nunokawa, E, Kodama, K, Matsuda, T, Yabuki, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-04-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selenomethionine incorporation into a protein by cell-free synthesis
J.STRUCT.FUNCT.GENOM., 2, 2001
|
|
1ZVJ
 
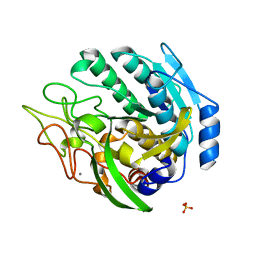 | | Structure of Kumamolisin-AS mutant, D164N | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Nakayama, T. | | Deposit date: | 2005-06-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Processing, catalytic activity and crystal structures of kumamolisin-As with an engineered active site.
Febs J., 273, 2006
|
|
8AJZ
 
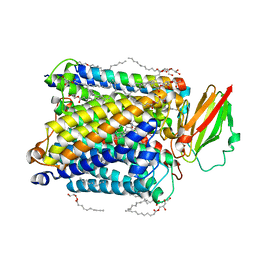 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase at 2 milliseconds after irradiation by a 532 nm laser | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Bosman, R, Dahl, P, Nango, E, Tanaka, R, Zoric, D, Svensson, E, Nakane, T, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
1ZVK
 
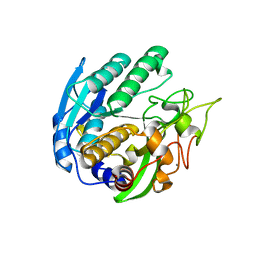 | | Structure of Double mutant, D164N, E78H of Kumamolisin-As | | Descriptor: | CALCIUM ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Nakayama, T. | | Deposit date: | 2005-06-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Processing, catalytic activity and crystal structures of kumamolisin-As with an engineered active site.
Febs J., 273, 2006
|
|
1GA6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PSCP (PSEUDOMONAS SERINE-CARBOXYL PROTEINASE) COMPLEXED WITH A FRAGMENT OF TYROSTATIN (THIS ENZYME RENAMED "SEDOLISIN" IN 2003) | | Descriptor: | ACETATE ION, CALCIUM ION, FRAGMENT OF TYROSTATIN, ... | | Authors: | Wlodawer, A, Li, M, Dauter, Z, Gustchina, A, Uchida, K. | | Deposit date: | 2000-11-29 | | Release date: | 2000-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Carboxyl proteinase from Pseudomonas defines a novel family of subtilisin-like enzymes.
Nat.Struct.Biol., 8, 2001
|
|
1GA4
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PSCP (PSEUDOMONAS SERINE-CARBOXYL PROTEINASE) COMPLEXED WITH INHIBITOR PSEUDOIODOTYROSTATIN (THIS ENZYME RENAMED "SEDOLISIN" IN 2003) | | Descriptor: | CALCIUM ION, GLYCEROL, PSEUDOIODOTYROSTATIN, ... | | Authors: | Wlodawer, A, Li, M, Dauter, Z, Gustchina, A, Uchida, K. | | Deposit date: | 2000-11-29 | | Release date: | 2000-12-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carboxyl proteinase from Pseudomonas defines a novel family of subtilisin-like enzymes.
Nat.Struct.Biol., 8, 2001
|
|
9C7D
 
 | |
9C79
 
 | |
9C7F
 
 | |
1GA1
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PSCP (PSEUDOMONAS SERINE-CARBOXYL PROTEINASE) COMPLEXED WITH A FRAGMENT OF IODOTYROSTATIN (THIS ENZYME RENAMED "SEDOLISIN" IN 2003) | | Descriptor: | CALCIUM ION, CHLORIDE ION, FRAGMENT OF IODOTYROSTATIN, ... | | Authors: | Dauter, Z, Li, M, Wlodawer, A. | | Deposit date: | 2000-11-29 | | Release date: | 2000-12-13 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Practical experience with the use of halides for phasing macromolecular structures: a powerful tool for structural genomics.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
