3Q3K
 
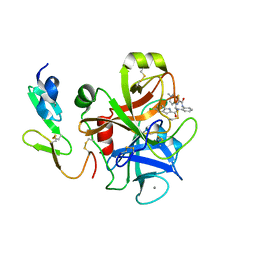 | | Factor Xa in complex with a phenylenediamine derivative | | Descriptor: | Activated factor Xa heavy chain, CALCIUM ION, Factor X light chain, ... | | Authors: | Suzuki, M, Imai, E. | | Deposit date: | 2010-12-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and SAR of novel ethylenediamine and phenylenediamine derivatives as factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1WTB
 
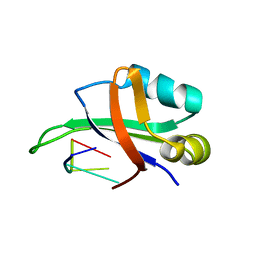 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D (AUF1) with telomere DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2004-11-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D
J.Biol.Chem., 280, 2005
|
|
7W9W
 
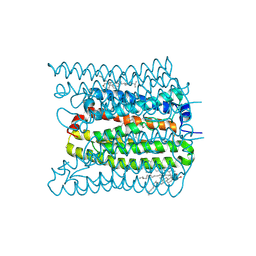 | | 2.02 angstrom cryo-EM structure of the pump-like channelrhodopsin ChRmine | | Descriptor: | CHOLESTEROL, ChRmine, PALMITIC ACID, ... | | Authors: | Kishi, K.E, Kim, Y, Fukuda, M, Yamashita, K, Deisseroth, K, Kato, H.E. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine.
Cell, 185, 2022
|
|
4IJ4
 
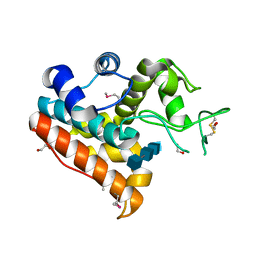 | | Crystal Structure of a Family GH19 chitinase from Bryum coronatum in complex with (GlcNAc)4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2012-12-21 | | Release date: | 2014-03-26 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a "loopless" GH19 chitinase in complex with chitin tetrasaccharide spanning the catalytic center.
Biochim.Biophys.Acta, 1844, 2014
|
|
3LLH
 
 | | Crystal structure of the first dsRBD of TAR RNA-binding protein 2 | | Descriptor: | MALONATE ION, RISC-loading complex subunit TARBP2 | | Authors: | Yamashita, S, Kawazoe, M, Takemoto, C, Sekine, S, Wakiyama, M, Yokoyama, S. | | Deposit date: | 2010-01-29 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The structures of dsRBDs of human TRBP
To be Published
|
|
8I2Z
 
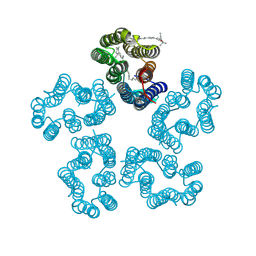 | | Cryo-EM structure of the zeaxanthin-bound kin4B8 | | Descriptor: | RETINAL, Xanthorhodopsin, Zeaxanthin | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2023-01-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
2D1J
 
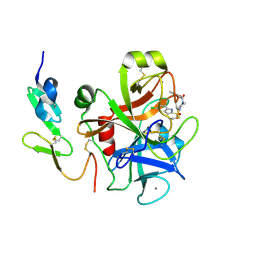 | | Factor Xa in complex with the inhibitor 2-[[4-[(5-chloroindol-2-yl)sulfonyl]piperazin-1-yl] carbonyl]thieno[3,2-b]pyridine n-oxide | | Descriptor: | 2-({4-[(5-CHLORO-1H-INDOL-2-YL)SULFONYL]PIPERAZIN-1-YL}CARBONYL)THIENO[3,2-B]PYRIDINE 4-OXIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Suzuki, M. | | Deposit date: | 2005-08-24 | | Release date: | 2006-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and biological activity of non-basic compounds as factor Xa inhibitors: SAR study of S1 and aryl binding sites.
Bioorg.Med.Chem., 13, 2005
|
|
7FBR
 
 | |
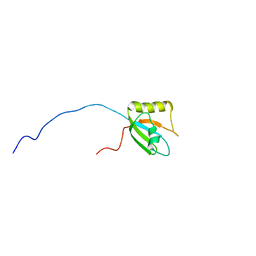
7FBV
 
 | |
2RSK
 
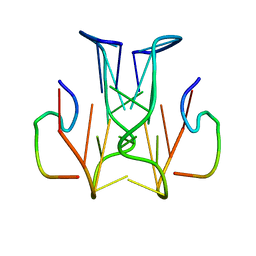 | | RNA aptamer against prion protein in complex with the partial binding peptide | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3'), partial binding peptide of Major prion protein | | Authors: | Mashima, T, Nishikawa, F, Kamatari, Y.O, Fujiwara, H, Nishikawa, S, Kuwata, K, Katahira, M. | | Deposit date: | 2012-03-08 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Anti-prion activity of an RNA aptamer and its structural basis
Nucleic Acids Res., 41, 2013
|
|
3WH1
 
 | | Crystal Structure of a Family GH19 Chitinase from Bryum coronatum in complex with (GlcNAc)4 at 1.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2013-08-21 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of a "loopless" GH19 chitinase in complex with chitin tetrasaccharide spanning the catalytic center.
Biochim.Biophys.Acta, 1844, 2014
|
|
1WU1
 
 | |
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8IU0
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Nakamura, S, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2023-03-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
1JQN
 
 | |
1JQO
 
 | |
2RPZ
 
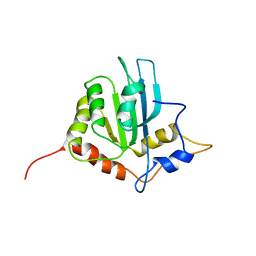 | | Solution structure of the monomeric form of mouse APOBEC2 | | Descriptor: | Probable C->U-editing enzyme APOBEC-2, ZINC ION | | Authors: | Hayashi, F, Nagata, T, Nagashima, T, Muto, Y, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-12-11 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monomeric form of mouse APOBEC2
To be Published
|
|
2RPA
 
 | | The solution structure of N-terminal domain of microtubule severing enzyme | | Descriptor: | Katanin p60 ATPase-containing subunit A1 | | Authors: | Iwaya, N, Kuwahara, Y, Unzai, S, Nagata, T, Tomii, K, Goda, N, Tochio, H, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2008-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A common substrate recognition mode conserved between katanin P60 and VPS4 governs microtubule severing and membrane skeleton reorganization
J.Biol.Chem., 285, 2010
|
|
