6B4D
 
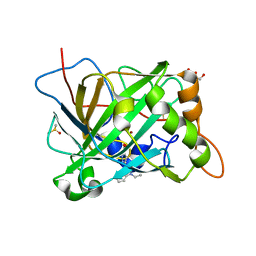 | | Crystal structure of human carbonic anhydrase II in complex with a heteroaryl-pyrazole carboxylic acid derivative. | | Descriptor: | 3-(1-ethyl-1H-indol-3-yl)-1-methyl-1H-pyrazole-5-carboxylic acid, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, Mahon, B.P, McKenna, R. | | Deposit date: | 2017-09-26 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.196 Å) | | Cite: | Exploring Heteroaryl-pyrazole Carboxylic Acids as Human Carbonic Anhydrase XII Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
6VJ3
 
 | | Carbonic Anhydrase II in complex with pyrimidine-based inhibitor | | Descriptor: | 2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-N-(4-sulfamoylphenyl)acetamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Lomelino, C.L, McKenna, R. | | Deposit date: | 2020-01-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inclusion of a 5-fluorouracil moiety in nitrogenous bases derivatives as human carbonic anhydrase IX and XII inhibitors produced a targeted action against MDA-MB-231 and T47D breast cancer cells.
Eur.J.Med.Chem., 190, 2020
|
|
6GEN
 
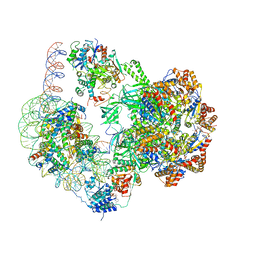 | | Chromatin remodeller-nucleosome complex at 4.5 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Willhoft, O, Chua, E.Y.D, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2018-04-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of the yeast SWR1-nucleosome complex.
Science, 362, 2018
|
|
6GEJ
 
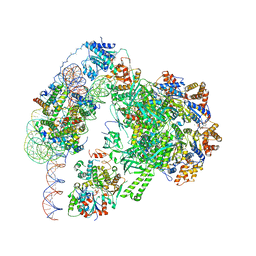 | | Chromatin remodeller-nucleosome complex at 3.6 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Willhoft, O, Chua, E.Y.D, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2018-04-26 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of the yeast SWR1-nucleosome complex.
Science, 362, 2018
|
|
4WG5
 
 | |
4WG3
 
 | |
4WG4
 
 | |
4ZGY
 
 | |
4ZGZ
 
 | |
5ZF6
 
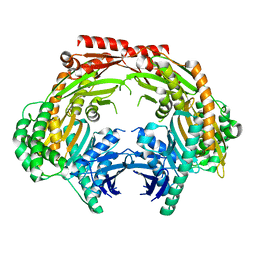 | | Crystal structure of the dimeric human PNPase | | Descriptor: | Polyribonucleotide nucleotidyltransferase 1, mitochondrial | | Authors: | Yuan, H.S, Golzarroshan, B. | | Deposit date: | 2018-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Crystal structure of dimeric human PNPase reveals why disease-linked mutants suffer from low RNA import and degradation activities.
Nucleic Acids Res., 46, 2018
|
|
6EBE
 
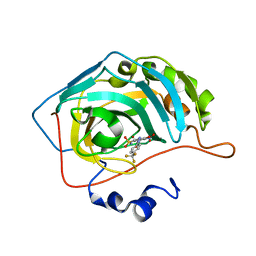 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 4-hydroxy-3-nitro-5-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
6EEA
 
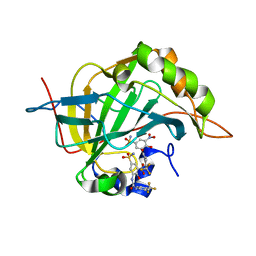 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 4-hydroxy-3-nitro-5-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
6EEH
 
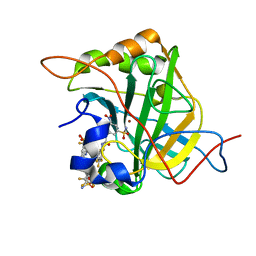 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | Carbonic anhydrase 2, N-[2-hydroxy-3-nitro-5-(nitrosulfonyl)phenyl]-N'-(pentafluorophenyl)urea, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-14 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
6EEO
 
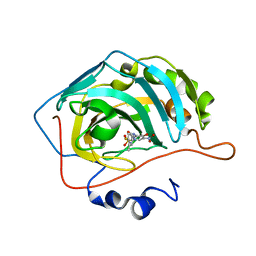 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 3-{[(4-fluoro-3-methylphenyl)carbamoyl]amino}-4-hydroxy-5-nitrobenzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
6EDA
 
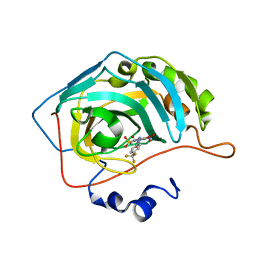 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 4-hydroxy-3-nitro-5-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-09 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
6ECZ
 
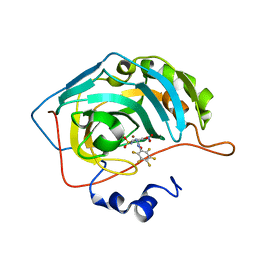 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | Carbonic anhydrase 2, N-[2-hydroxy-3-nitro-5-(nitrosulfonyl)phenyl]-N'-(pentafluorophenyl)urea, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
3NH2
 
 | |
3NH0
 
 | |
3NGZ
 
 | |
3NGY
 
 | | Crystal structure of RNase T (E92G mutant) | | Descriptor: | COBALT (II) ION, Ribonuclease T, his tag sequence | | Authors: | Hsiao, Y.-Y, Yuan, H.S. | | Deposit date: | 2010-06-14 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis for RNA trimming by RNase T in stable RNA 3'-end maturation
Nat.Chem.Biol., 7, 2011
|
|
3NH1
 
 | |
5XHV
 
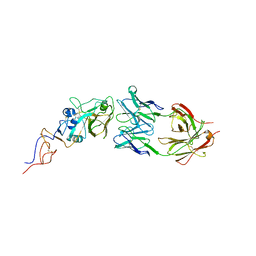 | | Crystal Structure Of Fab S40 In Complex With Influenza Hemagglutinin, HA1 subunit. | | Descriptor: | Hemagglutinin, S40 heavy chain, S40 light chain | | Authors: | Lee, C.C, Wang, A.H.J, Yu, C.M, Yang, A.S. | | Deposit date: | 2017-04-24 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | High throughput discovery of influenza virus neutralizing antibodies from phage-displayed synthetic antibody libraries.
Sci Rep, 7, 2017
|
|
4MXA
 
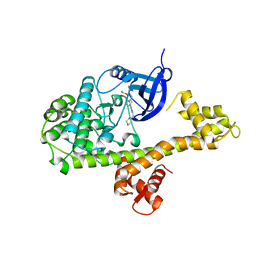 | | CDPK1 from Neospora caninum in complex with inhibitor RM-1-132 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-like domain protein kinase isoenzyme gamma, related | | Authors: | Merritt, E.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
4TZR
 
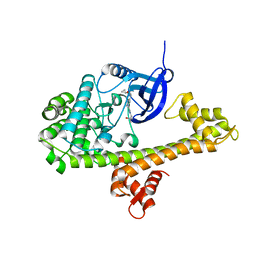 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with inhibitor UW1561 | | Descriptor: | 1-{4-amino-3-[2-(cyclopropyloxy)quinolin-6-yl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}-2-methylpropan-2-ol, Calmodulin-domain protein kinase 1 | | Authors: | Merritt, E.A. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an Orally Available and Central Nervous System (CNS) Penetrant Toxoplasma gondii Calcium-Dependent Protein Kinase 1 (TgCDPK1) Inhibitor with Minimal Human Ether-a-go-go-Related Gene (hERG) Activity for the Treatment of Toxoplasmosis.
J. Med. Chem., 59, 2016
|
|
3S5B
 
 | |
