5IFL
 
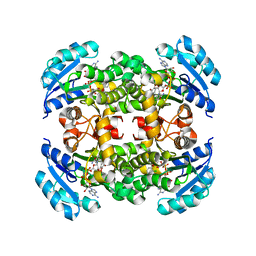 | | Crystal structure of B. pseudomallei FabI in complex with NAD and triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I7S
 
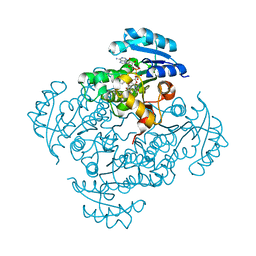 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT01 | | Descriptor: | 5-ETHYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I8W
 
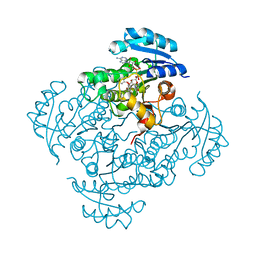 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT401 | | Descriptor: | 4-fluoro-5-hexyl-2-(2-methylphenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I50
 
 | | Structure of OmoMYC bound to double-stranded DNA | | Descriptor: | DNA (5'-D(P*CP*AP*CP*CP*CP*GP*GP*TP*CP*AP*CP*GP*TP*GP*GP*CP*CP*TP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*GP*GP*CP*CP*AP*CP*GP*TP*GP*AP*CP*CP*GP*GP*GP*TP*G)-3'), Myc proto-oncogene protein | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
5I7F
 
 | |
5I7E
 
 | |
5I9M
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT408 | | Descriptor: | 5-ethyl-4-fluoro-2-[(2-methylpyridin-3-yl)oxy]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5OBZ
 
 | | low resolution structure of the p34ct/p44ct minimal complex | | Descriptor: | Putative transcription factor, ZINC ION | | Authors: | Schoenwetter, E, Koelmel, W, Schmitt, D.R, Kuper, J, Kisker, C. | | Deposit date: | 2017-06-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
5NUS
 
 | | Structure of a minimal complex between p44 and p34 from Chaetomium thermophilum | | Descriptor: | ZINC ION, p34, p44 | | Authors: | Koelmel, W, Schoenwetter, E, Kuper, J, Schmitt, D.R, Kisker, C. | | Deposit date: | 2017-05-02 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
5I9L
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT404 | | Descriptor: | 2-(2-chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I9N
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT412 | | Descriptor: | 5-ethyl-4-fluoro-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
1Q51
 
 | | Crystal Structure of Mycobacterium tuberculosis MenB in Complex with Acetoacetyl-Coenzyme A, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | ACETOACETYL-COENZYME A, menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
1Q52
 
 | | Crystal Structure of Mycobacterium tuberculosis MenB, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
2EUA
 
 | | Structure and Mechanism of MenF, the Menaquinone-Specific Isochorismate Synthase from Escherichia Coli | | Descriptor: | D(-)-TARTARIC ACID, Menaquinone-specific isochorismate synthase | | Authors: | Kolappan, S, Kisker, C, Zwahlen, J, Zhou, R, Tonge, P.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lysine 190 is the catalytic base in MenF, the menaquinone-specific isochorismate synthase from Escherichia coli: implications for an enzyme family.
Biochemistry, 46, 2007
|
|
2FDC
 
 | |
2F4M
 
 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | Descriptor: | CHLORIDE ION, UV excision repair protein RAD23 homolog B, ZINC ION, ... | | Authors: | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2005-11-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
2F4O
 
 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | Descriptor: | CHLORIDE ION, PHQ-VAL-ALA-ASP-CF0, XP-C repair complementing complex 58 kDa protein, ... | | Authors: | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2005-11-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
5ONC
 
 | | Catabolism of the Cholesterol Side Chain in Mycobacterium tuberculosis is Controlled by a Redox-Sensitive Thiol Switch | | Descriptor: | CHLORIDE ION, Steroid 3-ketoacyl-CoA thiolase | | Authors: | Schaefer, C, Kuper, J, Sampson, N.S, Kisker, C. | | Deposit date: | 2017-08-03 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Catabolism of the Cholesterol Side Chain in Mycobacterium tuberculosis Is Controlled by a Redox-Sensitive Thiol Switch.
ACS Infect Dis, 3, 2017
|
|
1RGC
 
 | | THE COMPLEX BETWEEN RIBONUCLEASE T1 AND 3'-GUANYLIC ACID SUGGESTS GEOMETRY OF ENZYMATIC REACTION PATH. AN X-RAY STUDY | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Heydenreich, A, Koellner, G, Choe, H.W, Cordes, F, Kisker, C, Schindelin, H, Adamiak, R, Hahn, U, Saenger, W. | | Deposit date: | 1993-05-12 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The complex between ribonuclease T1 and 3'GMP suggests geometry of enzymic reaction path. An X-ray study.
Eur.J.Biochem., 218, 1993
|
|
2NRZ
 
 | |
2NRR
 
 | |
2NRX
 
 | | Crystal structure of the C-terminal half of UvrC, in the presence of sulfate molecules | | Descriptor: | GLYCEROL, SULFATE ION, UvrABC system protein C | | Authors: | Karakas, E, Truglio, J.J, Kisker, C. | | Deposit date: | 2006-11-02 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad.
Embo J., 26, 2007
|
|
1T5L
 
 | | Crystal structure of the DNA repair protein UvrB point mutant Y96A revealing a novel fold for domain 2 | | Descriptor: | UvrABC system protein B, ZINC ION | | Authors: | Truglio, J.J, Croteau, D.L, Skorvaga, M, DellaVecchia, M.J, Theis, K, Mandavilli, B.S, Van Houten, B, Kisker, C. | | Deposit date: | 2004-05-04 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactions between UvrA and UvrB: the role of UvrB's domain 2 in nucleotide excision repair
Embo J., 23, 2004
|
|
1Q9Y
 
 | | CRYSTAL STRUCTURE OF ENTEROBACTERIA PHAGE RB69 GP43 DNA POLYMERASE COMPLEXED WITH 8-OXOGUANOSINE CONTAINING DNA | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-AC(8-OXOGUANOSINE)GGTAAGCAGTCCGCG-3', 5'-GCGGACTGCTTAC(DIDEOXYCYTIDINE)-3', ... | | Authors: | Freisinger, E, Grollman, A.P, Miller, H, Kisker, C. | | Deposit date: | 2003-08-26 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lesion (in)tolerance reveals insights into DNA replication fidelity.
Embo J., 23, 2004
|
|
2NRW
 
 | |
