3CIR
 
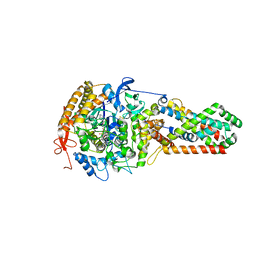 | | E. coli Quinol fumarate reductase FrdA T234A mutation | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2008-03-11 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | A threonine on the active site loop controls transition state formation in Escherichia coli respiratory complex II.
J.Biol.Chem., 283, 2008
|
|
6B58
 
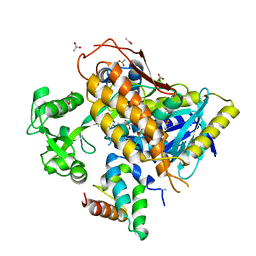 | | FrdA-SdhE assembly intermediate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, P, Iverson, T.M. | | Deposit date: | 2017-09-28 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Crystal structure of an assembly intermediate of respiratory Complex II.
Nat Commun, 9, 2018
|
|
8T8P
 
 | |
8T8O
 
 | |
8SV2
 
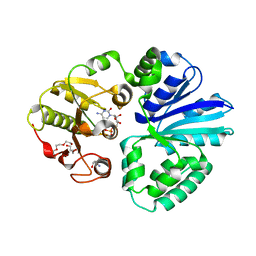 | | Pasteurella multocida alpha2,3/2,6 sialyltransferase D141N bound to CMP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-2,3/2,6-sialyltransferase/sialidase, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Stubbs, H.E, Iverson, T.M. | | Deposit date: | 2023-05-15 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Pasteurella multocida alpha2,3/2,6 sialyltransferase D141N bound to CMP
To Be Published
|
|
6VAX
 
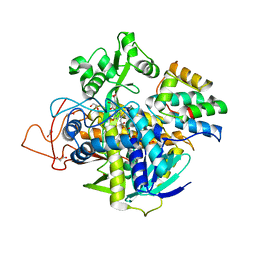 | | Crystal structure of human SDHA-SDHAF2 assembly intermediate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Sharma, P, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2019-12-18 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The roles of SDHAF2 and dicarboxylate in covalent flavinylation of SDHA, the human complex II flavoprotein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8ST6
 
 | |
8ST5
 
 | |
6VT2
 
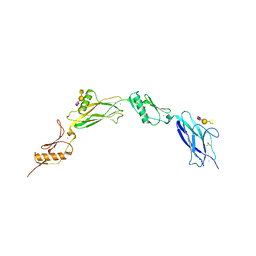 | |
6VU6
 
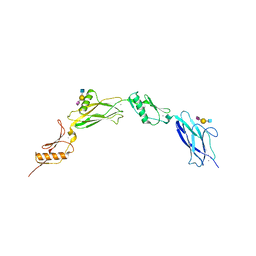 | |
6VS7
 
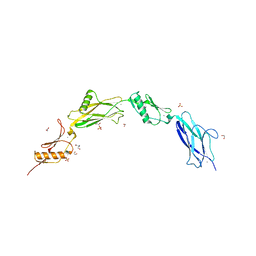 | | Sialic acid binding region of Streptococcus Sanguinis SK1 adhesin | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adhesin, ... | | Authors: | Stubbs, H.E, Iverson, T.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tandem sialoglycan-binding modules in a Streptococcus sanguinis serine-rich repeat adhesin create target dependent avidity effects.
J.Biol.Chem., 295, 2020
|
|
8SHH
 
 | | Crystal structure of EvdS6 decarboxylase in ligand free state | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-glucose 4,6-dehydratase | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
8SK0
 
 | | Crystal structure of EvdS6 decarboxylase in ligand bound state | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
5EQ4
 
 | | Crystal structure of the SrpA adhesin R347E mutant from Streptococcus sanguinis | | Descriptor: | ACETATE ION, CALCIUM ION, Platelet-binding glycoprotein | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
5EQ2
 
 | | Crystal Structure of the SrpA Adhesin from Streptococcus sanguinis | | Descriptor: | ACETATE ION, CALCIUM ION, Platelet-binding glycoprotein | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
5EQ3
 
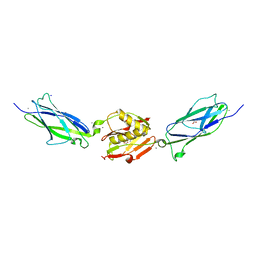 | | Crystal structure of the SrpA adhesin from Streptococcus sanguinis with a sialyl galactose disaccharide bound | | Descriptor: | ACETATE ION, CALCIUM ION, N-glycolyl-alpha-neuraminic acid-(2-3)-methyl beta-D-galactopyranoside, ... | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
6EC3
 
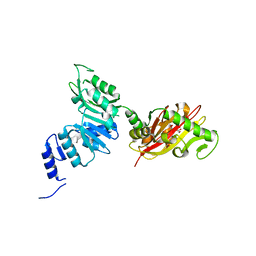 | | Crystal Structure of EvdMO1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Methyltransferase domain-containing protein, NICKEL (II) ION | | Authors: | McCulloch, K.M, Iverson, T.M, Starbird, C.A, Perry, N.A, Chen, Q, Berndt, S, Yamakawa, I, Loukachevitch, L.V. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.
Biochemistry, 57, 2018
|
|
3P4P
 
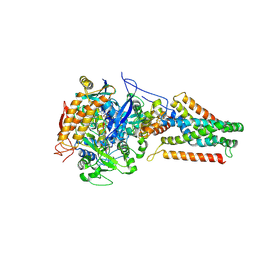 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with fumarate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andr ll, J, Luna-Ch vez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
3P4Q
 
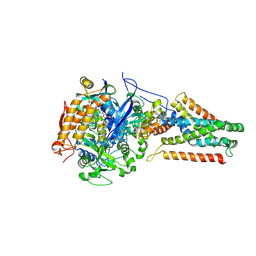 | | Crystal structure of Menaquinol:oxidoreductase in complex with oxaloacetate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
3P4R
 
 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with glutarate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
3P4S
 
 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with a 3-nitropropionate adduct | | Descriptor: | 3-NITROPROPANOIC ACID, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
4KX6
 
 | | Plasticity of the quinone-binding site of the complex II homolog quinol:fumarate reductase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Singh, P.K, Sarwar, M, Maklashina, E, Kotlyar, V, Rajagukguk, S, Tomasiak, T.M, Cecchini, G, Iverson, T.M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plasticity of the Quinone-binding Site of the Complex II Homolog Quinol:Fumarate Reductase.
J.Biol.Chem., 288, 2013
|
|
4N0E
 
 | | Crystal structure of the K345L variant of the Gi alpha1 subunit bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, SULFATE ION | | Authors: | Thaker, T.M, Preininger, A.M, Sarwar, M, Hamm, H.E, Iverson, T.M. | | Deposit date: | 2013-10-01 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Transient Interaction between the Phosphate Binding Loop and Switch I Contributes to the Allosteric Network between Receptor and Nucleotide in G alpha i1.
J.Biol.Chem., 289, 2014
|
|
6NBS
 
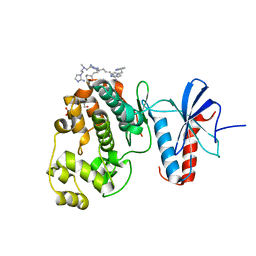 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
6NMW
 
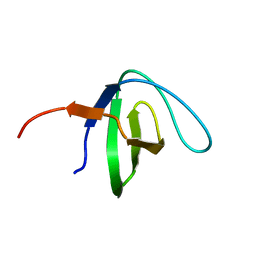 | |
