4ZXF
 
 | | Crystal Structure of a Soluble Variant of Monoglyceride Lipase from Saccharomyces Cerevisiae in Complex with a Substrate Analog | | Descriptor: | 1-{3-[(R)-hydroxy(octadecyloxy)phosphoryl]propyl}triaza-1,2-dien-2-ium, Monoglyceride lipase, NITRATE ION, ... | | Authors: | Aschauer, P, Lichtenegger, J, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
3GFS
 
 | | Structure of YhdA, K109D/D137K variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADPH-azoreductase | | Authors: | Staunig, N, Gruber, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase
Febs J., 276, 2009
|
|
5MP4
 
 | | The structure of Pst2p from Saccharomyces cerevisiae | | Descriptor: | PHOSPHATE ION, Protoplast secreted protein 2 | | Authors: | Hromic, A, Gruber, K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure, biochemical and kinetic properties of recombinant Pst2p from Saccharomyces cerevisiae, a FMN-dependent NAD(P)H:quinone oxidoreductase.
Biochim. Biophys. Acta, 1865, 2017
|
|
5A2G
 
 | | An esterase from anaerobic Clostridium hathewayi can hydrolyze aliphatic aromatic polyesters | | Descriptor: | CARBOXYLIC ESTER HYDROLASE, PHOSPHATE ION | | Authors: | Hromic, A, Pavkov Keller, T, Steinkellner, G, Gruber, K, Perz, V, Baumschlager, A, Bleymaier, K, Zitzenbacher, S, Zankel, A, Mayrhofer, C, Sinkel, C, Kueper, U, Schlegel, K.A, Ribitsch, D, Guebitz, G.M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | An Esterase from Anaerobic Clostridium Hathewayi Can Hydrolyze Aliphatic-Aromatic Polyesters.
Environ.Sci.Tech., 50, 2016
|
|
5A4K
 
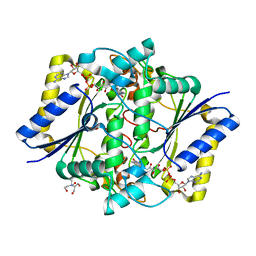 | | Crystal structure of the R139W variant of human NAD(P)H:quinone oxidoreductase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Lienhart, W.D, Strandback, E, Gudipati, V, Uhl, M.K, Rantase, D.M, Zangger, K, Gruber, K, Macheroux, P. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Catalytic competence, structure and stability of the cancer-associated R139W variant of the human NAD(P)H:quinone oxidoreductase 1 (NQO1).
FEBS J., 284, 2017
|
|
5L46
 
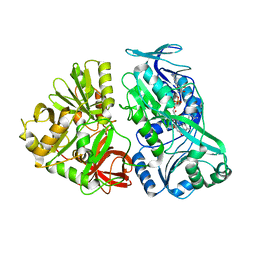 | |
5LUI
 
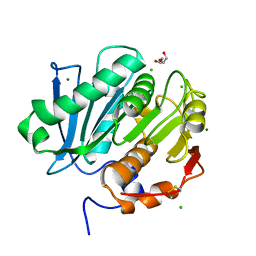 | | Structure of cutinase 1 from Thermobifida cellulosilytica | | Descriptor: | CHLORIDE ION, Cutinase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hromic, A, Lyskowski, A, Gruber, K. | | Deposit date: | 2016-09-08 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small cause, large effect: Structural characterization of cutinases from Thermobifida cellulosilytica.
Biotechnol. Bioeng., 114, 2017
|
|
5LUK
 
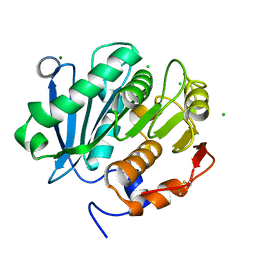 | | Structure of a double variant of cutinase 2 from Thermobifida cellulosilytica | | Descriptor: | CHLORIDE ION, Cutinase 2, MAGNESIUM ION | | Authors: | Hromic, A, Lyskowski, A, Gruber, K. | | Deposit date: | 2016-09-09 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Small cause, large effect: Structural characterization of cutinases from Thermobifida cellulosilytica.
Biotechnol. Bioeng., 114, 2017
|
|
5LUJ
 
 | |
5LUL
 
 | | Structure of a triple variant of cutinase 2 from Thermobifida cellulosilytica | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cutinase 2 | | Authors: | Hromic, A, Lyskowski, A, Gruber, K. | | Deposit date: | 2016-09-09 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small cause, large effect: Structural characterization of cutinases from Thermobifida cellulosilytica.
Biotechnol. Bioeng., 114, 2017
|
|
4JIP
 
 | |
4JIC
 
 | | Glycerol Trinitrate Reductase NerA from Agrobacterium radiobacter | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, GTN Reductase, ... | | Authors: | Oberdorfer, G, Gruber, K. | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of Glycerol Trinitrate Reductase NerA from Agrobacterium radiobacter Reveals the Molecular Reason for Nitro- and Ene-Reductase Activity in OYE Homologues.
Chembiochem, 14, 2013
|
|
4JIQ
 
 | |
3GDN
 
 | | Almond hydroxynitrile lyase in complex with benzaldehyde | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dreveny, I, Gruber, K, Kratky, C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
Biochemistry, 48, 2009
|
|
1ZCH
 
 | | Structure of the hypothetical oxidoreductase YcnD from Bacillus subtilis | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Morokutti, A, Lyskowski, A, Sollner, S, Pointner, E, Fitzpatrick, T.B, Kratky, C, Gruber, K, Macheroux, P. | | Deposit date: | 2005-04-12 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Function of YcnD from Bacillus subtilis, a Flavin-Containing Oxidoreductase(,).
Biochemistry, 44, 2005
|
|
5AH1
 
 | | Structure of EstA from Clostridium botulinum | | Descriptor: | POTASSIUM ION, TRIACYLGLYCEROL LIPASE, ZINC ION | | Authors: | Pairitsch, A, Lyskowski, A, Hromic, A, Steinkellner, G, Gruber, K, Perz, V, Baumschlager, A, Bleymaier, K, Zitzenbacher, S, Sinkel, C, Kueper, U, Ribitsch, D, Guebitz, G.M. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hydrolysis of Synthetic Polyesters by Clostridium Botulinum Esterases.
Biotechnol.Bioeng., 113, 2016
|
|
3GFR
 
 | | Structure of YhdA, D137L variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADPH-azoreductase | | Authors: | Staunig, N, Gruber, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase
Febs J., 276, 2009
|
|
3GFQ
 
 | | Structure of YhdA, K109L variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADPH-azoreductase | | Authors: | Staunig, N, Gruber, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.996 Å) | | Cite: | A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase
Febs J., 276, 2009
|
|
5AH0
 
 | | STRUCTURE OF LIPASE 1 FROM PELOSINUS FERMENTANS | | Descriptor: | DI(HYDROXYETHYL)ETHER, LIPASE, POTASSIUM ION, ... | | Authors: | Hromic, A, Gruber, K, Biundo, A, Ribitsch, D, Quartinello, F, Perz, V, Arrell, M.S, Kalman, F, Guebitz, G.M. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of a Poly(Butylene Adipate-Co-Terephthalate)-Hydrolyzing Lipase from Pelosinus Fermentans.
Appl.Microbiol.Biotechnol., 100, 2016
|
|
4D7K
 
 | |
4D1Y
 
 | | Crystal structure of a putative protease from Bacteroides thetaiotaomicron. | | Descriptor: | PUTATIVE PROTEASE I, RIBOFLAVIN, ZINC ION | | Authors: | Knaus, T, Uhl, M.K, Monschein, S, Moratti, S, Gruber, K, Macheroux, P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Stability of an Unusual Zinc-Binding Protein from Bacteroides Thetaiotaomicron.
Biochim.Biophys.Acta, 1844, 2014
|
|
4EC3
 
 | | Structure of berberine bridge enzyme, H174A variant in complex with (S)-reticuline | | Descriptor: | (S)-reticuline, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Winkler, A, Macheroux, P, Gruber, K. | | Deposit date: | 2012-03-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6501 Å) | | Cite: | Catalytic and structural role of a conserved active site histidine in berberine bridge enzyme.
Biochemistry, 51, 2012
|
|
5MG5
 
 | | A multi-component acyltransferase PhlABC from Pseudomonas protegens soaked with the monoacetylphloroglucinol (MAPG) | | Descriptor: | 2,4-diacetylphloroglucinol biosynthesis protein, 2,4-diacetylphloroglucinol biosynthesis protein PhlC, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Pavkov-Keller, T, Schmidt, N.G, Kroutil, W, Gruber, K. | | Deposit date: | 2016-11-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Structure and Catalytic Mechanism of a Bacterial Friedel-Crafts Acylase.
Chembiochem, 20, 2019
|
|
5M3K
 
 | | A multi-component acyltransferase PhlABC from Pseudomonas protegens | | Descriptor: | 2,4-diacetylphloroglucinol biosynthesis protein PhlB, 2,4-diacetylphloroglucinol biosynthesis protein PhlC, PhlA, ... | | Authors: | Pavkov-Keller, T, Schmidt, N.G, Kroutil, W, Gruber, K. | | Deposit date: | 2016-10-15 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure and Catalytic Mechanism of a Bacterial Friedel-Crafts Acylase.
Chembiochem, 20, 2019
|
|
3GDP
 
 | | Hydroxynitrile lyase from almond, monoclinic crystal form | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dreveny, I, Gruber, K, Kratky, C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
Biochemistry, 48, 2009
|
|
